- Title
-
Exploring Zebrafish Larvae as a COVID-19 Model: Probable Abortive SARS-CoV-2 Replication in the Swim Bladder
- Authors
- Laghi, V., Rezelj, V., Boucontet, L., Frétaud, M., Da Costa, B., Boudinot, P., Salinas, I., Lutfalla, G., Vignuzzi, M., Levraud, J.P.
- Source
- Full text @ Front Cell Infect Microbiol
|
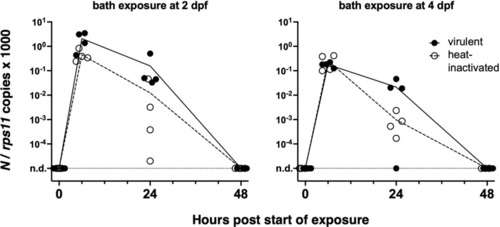
Bath exposure of zebrafish larvae to SARS-CoV-2. Kinetics of qRT-PCR measurements of polyadenylated viral N copies; each point corresponds to an individual larva. n.d., not detected. |
|
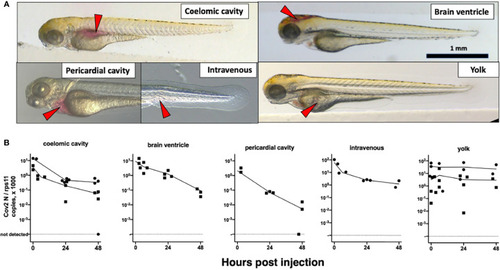
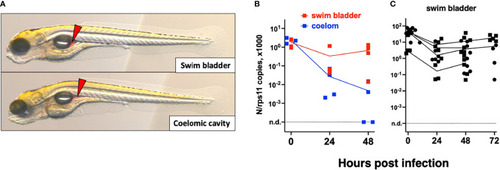
Microinjection of SARS-CoV-2 to 3-dpf wild-type larvae. |
|
Microinjection of SARS-CoV-2 to 4-dpf larvae. |
|
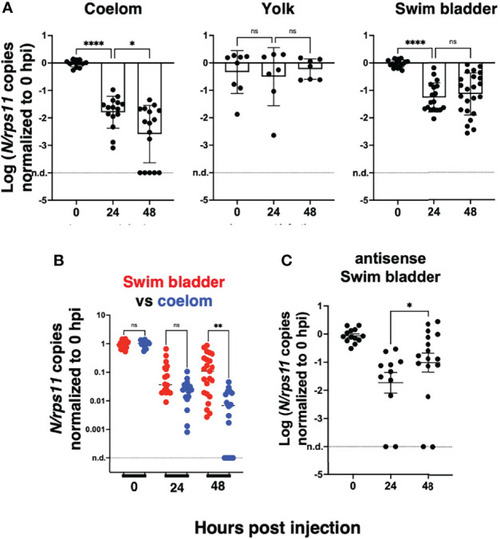
Statistical analysis of viral transcript quantifications, after normalization to the means of values measured at 0 hpi for each experiment. |
|
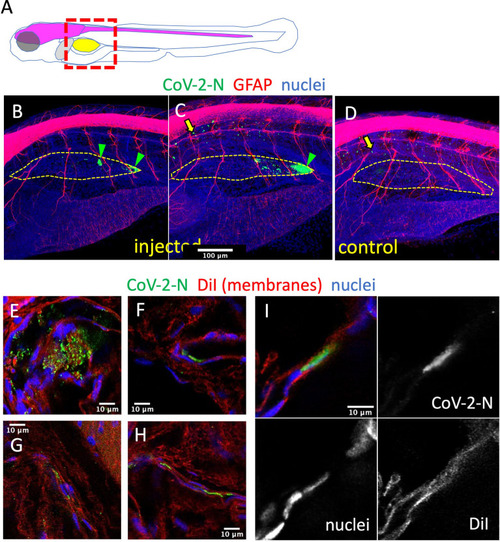
Immunodetection of infected cells in the swim bladder. EXPRESSION / LABELING:
|
|
Testing SARS-CoV-2 variants. qRT-PCR analysis of larvae at various times after injection of 2 nl of virus suspension in the swim bladder. Dotted lines separate independent experiments. |
|
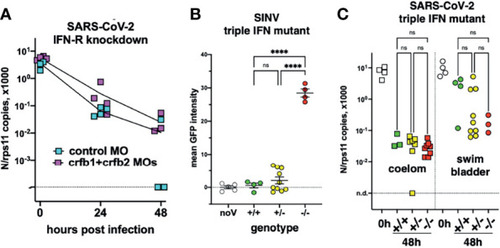
Viral infection in IFN-defective larvae. |
|
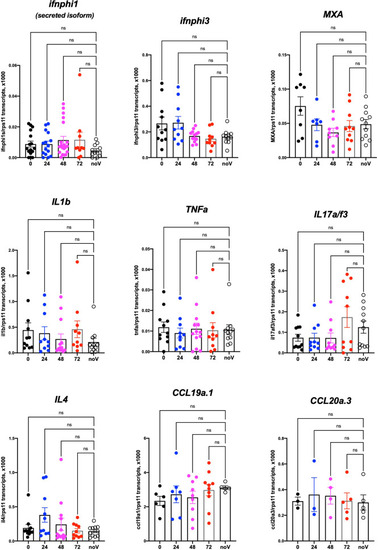
Host response after SARS-CoV-2 injection in the swim bladder. qRT-PCR, pool of 4 independent experiments (except for ccl19a.1 and ccl20a.3, 3 and 2 experiments respectively). Numbers on the X-axis refer to hours postinjection; noV (for “no Virus”): pooled uninjected negative controls, age-matched to 24, 48, or 72 hpi. One-way ANOVA analysis. ns, not significant. |
|
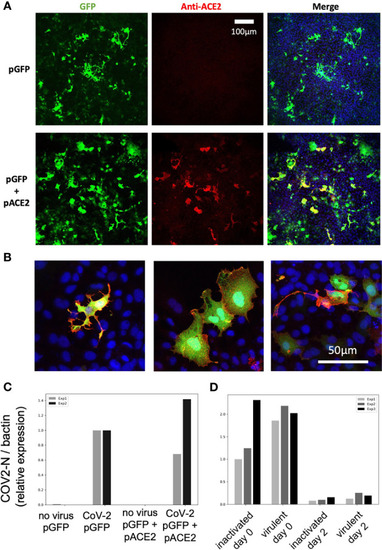
Overexpression of hACE2-mCherry by plasmid injection at the 1-cell stage. |
|
Injection of 3-dpf hACE2-mCherry mosaic larvae. Quantification of sense N transcripts in individual hACE2-mCherry mosaic larvae injected in coelomic cavity (left; one experiment with hACE2-mCherry, one with hACE2-2A-mCherry) or brain ventricle (right; with hACE2-2A-mCherry) by qRT-PCR. |
|
SARS-CoV-2 does not replicate on EPC cells transfected with human ACE2. |