- Title
-
The H3K27 demethylase controls the lateral line embryogenesis of zebrafish
- Authors
- Tang, D., Lu, Y., Zuo, N., Yan, R., Wu, C., Wu, L., Liu, S., He, Y.
- Source
- Full text @ Cell Biol. Toxicol.
|
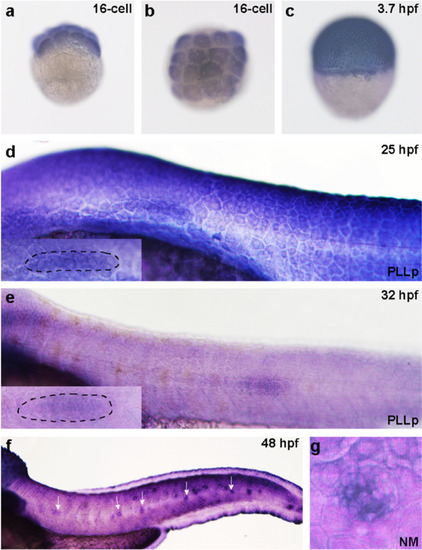
The expression pattern of |
|
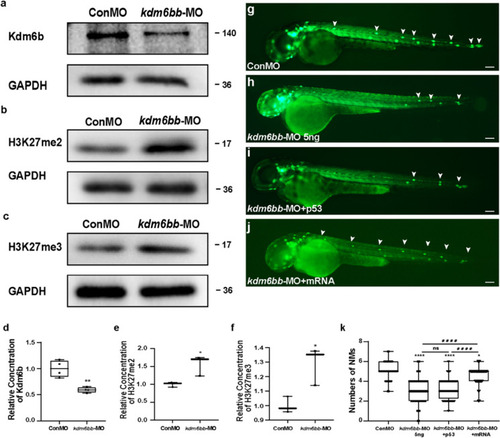
Kdm6b is required for cell migration and neuromast deposition in zebrafish posterior lateral lines. |
|
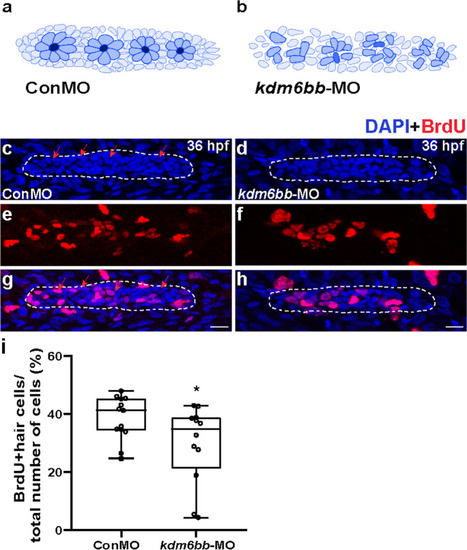
The downregulation of Kdm6b represses cell proliferation and disrupts rosette assembly during primordium migration. |
|
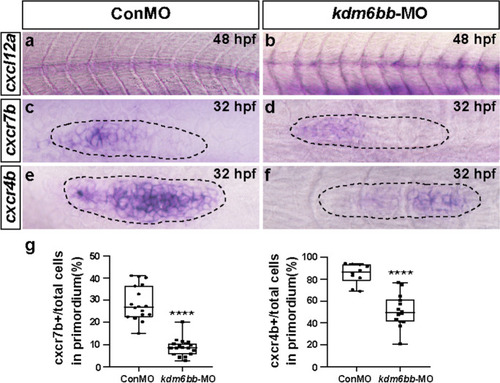
The chemokine signaling pathway is disrupted by |
|
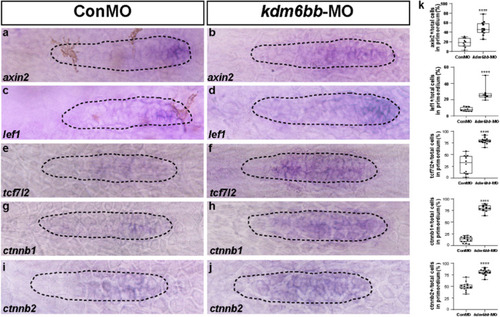
Knockdown of Kdm6b upregulates the Wnt signaling pathway. |
|
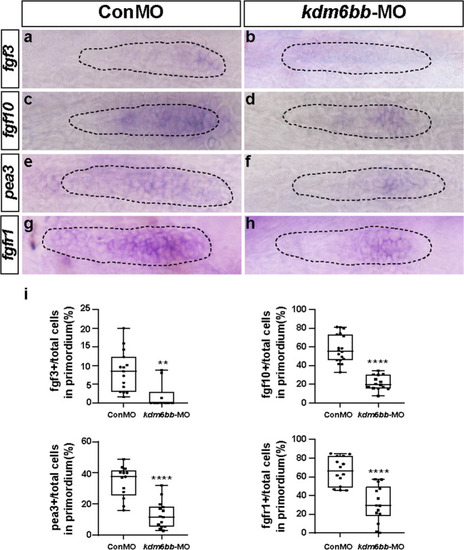
Kdm6b-depletion represses Fgf signaling in zebrafish primordium. |