- Title
-
The E3 ubiquitin ligase component, Cereblon, is an evolutionarily conserved regulator of Wnt signaling
- Authors
- Shen, C., Nayak, A., Neitzel, L.R., Adams, A.A., Silver-Isenstadt, M., Sawyer, L.M., Benchabane, H., Wang, H., Bunnag, N., Li, B., Wynn, D.T., Yang, F., Garcia-Contreras, M., Williams, C.H., Dakshanamurthy, S., Hong, C.C., Ayad, N.G., Capobianco, A.J., Ahmed, Y., Lee, E., Robbins, D.J.
- Source
- Full text @ Nat. Commun.
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
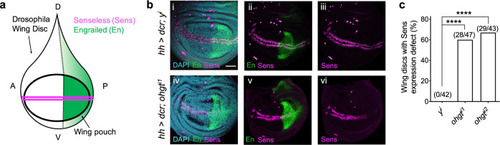
EXPRESSION / LABELING:
PHENOTYPE:
|
|
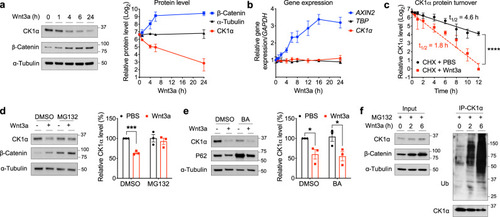
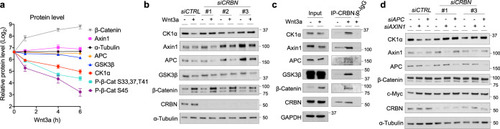
Upon stimulation of Wnt signaling, CRBN is recruited to the β-Catenin destruction complex and targets CK1α for degradation, further promoting Wnt pathway activation. |

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. EXPRESSION / LABELING:
PHENOTYPE:
|

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. |