- Title
-
Identification of Rhythmically Expressed LncRNAs in the Zebrafish Pineal Gland and Testis
- Authors
- Mishra, S.K., Liu, T., Wang, H.
- Source
- Full text @ Int. J. Mol. Sci.
|
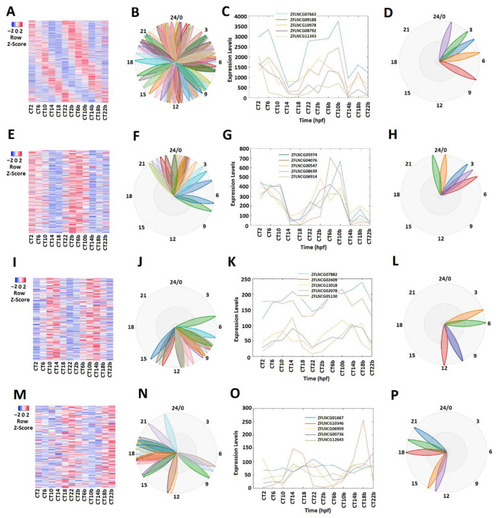
Expression profile analysis of morning (CT 2 and CT 6), evening (CT10 and CT14), and night (CT18 and CT22) rhythmically expressed pineal gland lncRNAs. Analysis of all 586 rhythmically expressed pineal gland lncRNAs (A–D): heat map (A) and phases (B) of all 586 rhythmically expressed pineal gland lncRNAs; expression profiles (C) and phases (D) of representative lncRNAs. Analysis of 241 pineal gland morning lncRNAs (E–H): heat map (E) and phases (F) of 241 pineal gland morning lncRNAs; expression profiles (G) and phases (H) of representative pineal gland morning lncRNAs. Analysis of 196 pineal gland evening lncRNAs (I–L): heat map (I) and phases (J) of 189 pineal gland evening lncRNAs; expression profiles (K) and phases (L) of representative pineal gland evening lncRNAs. Analysis of 149 pineal gland night lncRNAs (M–P): heat map (M) and phases (N) of 217 pineal gland night lncRNAs; expression profiles (O) and phases (P) of representative pineal gland night lncRNAs. |
|
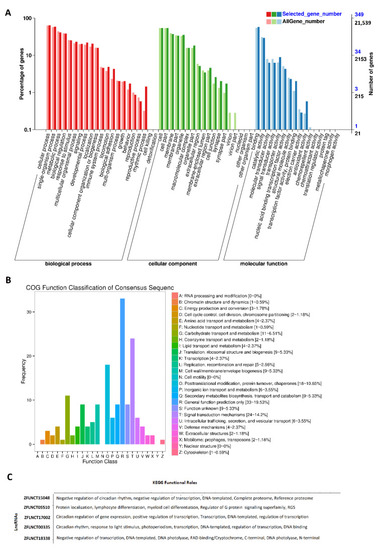
GO, COG, and KEGG analyses of rhythmically expressed pineal gland lncRNAs: GO annotation revealed the percentage of genes involved in different biological processes, cellular components (e.g., cell membrane part, macromolecular complex, and synapse), and molecular functions (A), COG functional classification of lncRNAs (B), and KEGG functional roles of the representative lncRNAs (C). |
|
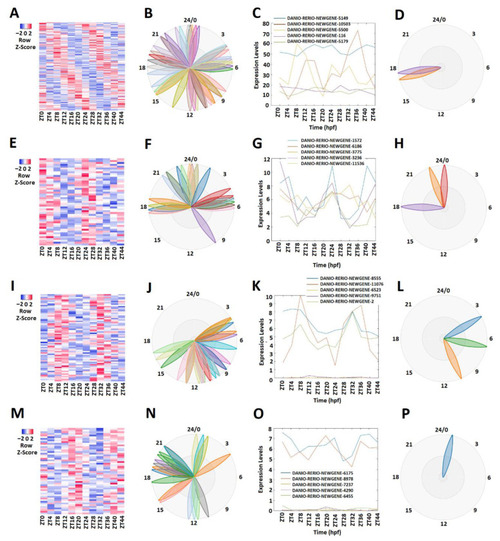
Expression profile analysis of morning (ZT 0 and ZT4), evening (ZT8 and ZT12), and night (ZT 16 and ZT 20) rhythmically expressed testis lncRNAs. Analysis of all 165 rhythmically expressed testis lncRNAs (A–D): heat map (A) and phases (B) of all 165 rhythmically expressed testis lncRNAs; expression profiles (C) and phases (D) of representative lncRNAs. Analysis of 47 testis morning lncRNAs (E–H): heat map (E) and phases (F) of 47 testis morning lncRNAs; expression profiles (G) and phases (H) of representative testis morning lncRNAs. Analysis of 66 testis evening lncRNAs (I–L): heat map (I) and phases (J) of 66 testis evening lncRNAs; expression profiles (K) and phases (L) of representative testis evening lncRNAs. Analysis of 52 testis lncRNAs (M–P): heat map (M) and phases (N) of 52 testis night lncRNAs; expression profiles (O) and phases (P) of representative testis night lncRNAs. |
|
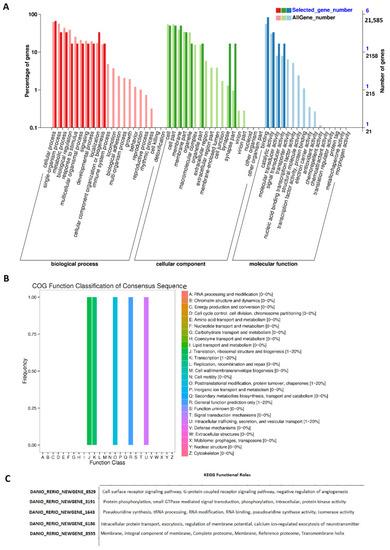
GO, COG, and KEGG analysis of rhythmically expressed testis lncRNAs: GO annotation revealed the percentage of genes involved in different biological processes, cellular components (e.g., cell membrane part, macromolecular complex, and synapse), and molecular functions (A), COG functional classification of lncRNAs (B), and KEGG functional roles of the representative lncRNAs (C). |
|
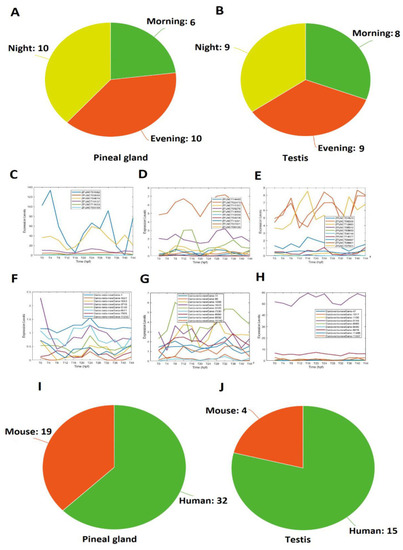
Overlapping lncRNAs between the pineal gland and testis ( |
|
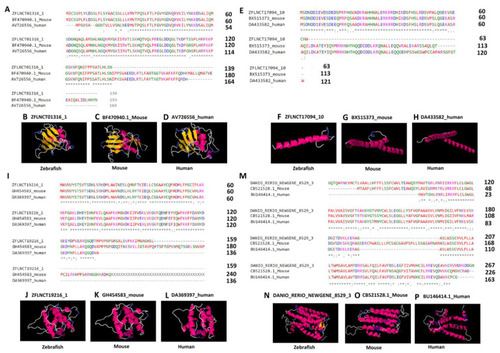
Multiple sequence alignment and three-dimensional structures predicted by (PS)2-v2: protein structure prediction server for the highly conserved lncRNA-encoded peptides among zebrafish, humans, and mice. Pineal gland lncRNA-encoded peptides (A–L) and testis lncRNA-encoded peptides (M–P). The asterisk symbols (A,E,I,M) depict the identical amino acids among the peptides encoded by zebrafish lncRNAs (ZFLNCT01316_1, ZFLNCT17094_10, ZFLNCT19216_1, and DANIO_RERIO_NEWGENE_8529_1), as well as their corresponding mouse orthologs (BF470940.1_Mouse, BX515373_mouse, GH454583_mouse, and CB521528.1_Mouse) and human orthologs (AV726556_human, DA433582_human, DA369397_human, and BU146414.1_Human). The 3D models of the peptides (B–D,F–H,J–L,N–P) represent the conservation of α-helix (pink or purple motif structure), β-strand (yellow layered band), and random coils (white or blue thread) among zebrafish, humans, and mice with known domains from the Protein Data Bank, such as, B (2adcA), C (2adcA), D (2adcA), F (1nkpB), G (1nkpB), H (1nkpB), J (1wyoA), K (1wyoA), L (1wyoA), N (1u19A), O (2vt4B), and P (1xioA). |