- Title
-
Whole brain functional recordings at cellular resolution in zebrafish larvae with 3D scanning multiphoton microscopy
- Authors
- Bruzzone, M., Chiarello, E., Albanesi, M., Miletto Petrazzini, M.E., Megighian, A., Lodovichi, C., Dal Maschio, M.
- Source
- Full text @ Sci. Rep.
|
The layout of the optical configuration and the properties of the ETL. ( |
|
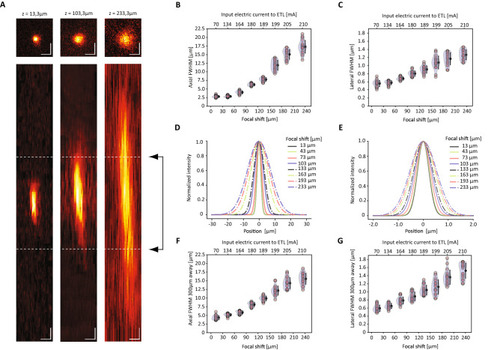
Optical performances of the system. (A) Average intensity projections (upper row with top-view XY, lower row with side view ZX) of the point spread function (PSF) measured at 920 nm on 0.2 μm fluorescent beads at three different settings of the ETL tuning range, z = 13.3 μm, z = 103.3 μm and z = 233.3 μm. Highlighted with dashed lines is the typical cell size (6 μm). Scale bar = 1 μm. (B,C) Axial and lateral FWHM of the PSF as function of the ETL defocus and of the corresponding ETL driving current, for beads measured within 50 μm from the center of the FOV. (D,E) Axial and lateral intensity profile of the PSF for beads measured within 50 μm from the center, corresponding to eight different settings of the ETL. (F,G) Axial and lateral FWHM of the PSF as function of the ETL defocus and of the corresponding ETL driving current, for beads measured at 300 μm from the center of the FOV. |
|
Multiplane calcium imaging of the RGC arborization fields. ( |
|
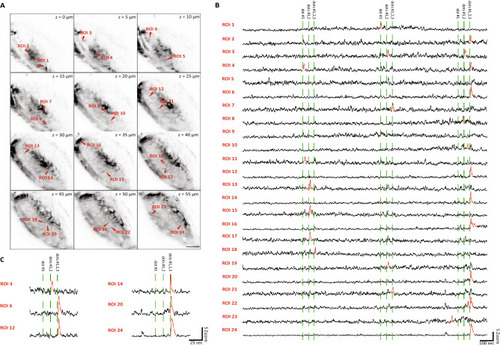
Whole brain calcium imaging. ( |
|
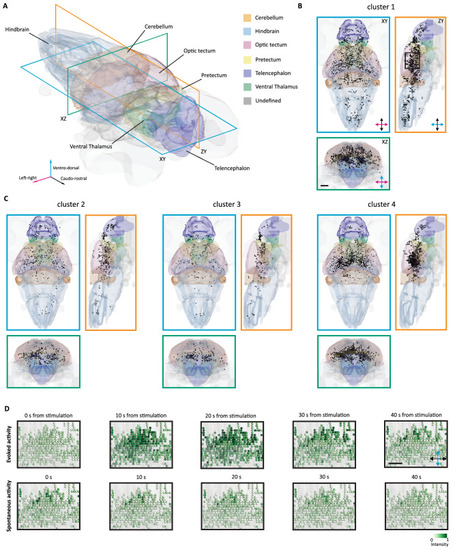
Anatomical representation of the identified clusters of visually responsive cells. (A) Three-dimensional representation of the brain of the larva with some of the most prominent region indicated. Three sectioning planes are overlaid to indicate the corresponding viewports for the following orthoviews. (B) Horizontal (cerulean), sagittal (orange) and coronal (bluish-green) views of the brain, with the projection of the stimulus-responsive cells belonging to the cluster 1. The black box indicates the area of the inset in D. Scale bar = 100 μm. (C) Horizontal, sagittal and coronal views of the brain, with the projection of the stimulus-responsive cells belonging to the cluster 2, 3 and 4. (D) Field of view extracted from the longitudinal plane in (B). In the top row, evoked activity of visually responsive cells is reported for 40 s from the onset of the stimulus. In the bottom row, spontaneous activity of the same cells is reported for 40 s. Scale bar = 50 μm. |