- Title
-
Upregulation of the pathogenic transcription factor SPI1/PU.1 in tuberous sclerosis complex and focal cortical dysplasia by oxidative stress
- Authors
- Zimmer, T.S., Korotkov, A., Zwakenberg, S., Jansen, F.E., Zwartkruis, F.J.T., Rensing, N.R., Wong, M., Mühlebner, A., van Vliet, E.A., Aronica, E., Mills, J.D.
- Source
- Full text @ Brain Pathol
|
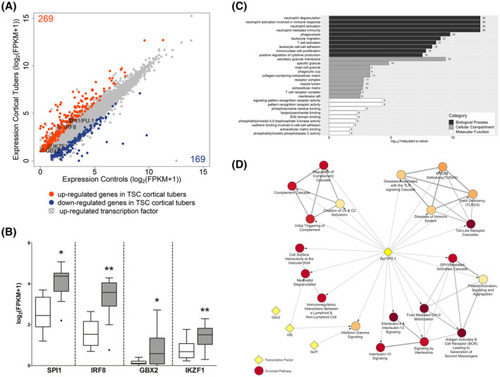
Identification of SPI1, IRF8, GBX2, and IKZF1 as novel transcription factors in TSC cortical tuber. Differential expression analysis of cortical tuber tissue versus age‐matched cortical control samples derived from autopsy revealed 269 upregulated (orange) and 169 downregulated (blue) genes. Four potentially important transcriptional regulators were identified from the upregulated genes (cross circle) (A). Comparison of log2FPKM values from cortical tuber tissue versus age‐matched cortical control tissue from autopsy (B). GO enrichment analysis of the top 10 enriched pathways of 139 genes predicted to be targeted by SPI1/PU.1 identified predominantly immune system‐associated GO terms (C). Reactome pathway enrichment analysis of the upregulated genes from RNA Seq revealed SPI1/PU.1 to target multiple genes in the top 20 most enriched pathway. IRF8 targeted 2 pathways, while GBX2 and IKZF1 targeted only 1 pathway. All pathways shown here have an adjusted |
|
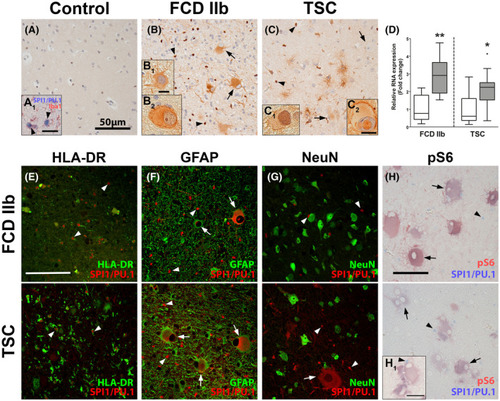
SPI1/PU.1 is highly expressed in malformed cells in TSC and FCD 2b lesions. Autopsy control cortex showed SPI1/PU.1 expression solely in microglia (A, A1). In TSC and FCD 2b expression of SPI1/PU.1 was found in microglia (arrowheads) but also malformed cells and dysmorphic neurons (arrows) (B, B1, C). Moreover, SPI1/PU.1 expression could also be found in the nuclei of malformed cells (B2, C1, C2). Analysis of RNA expression in tissue from resected TSC tuber and FCD 2b lesion of an independent cohort showed higher expression of |
|
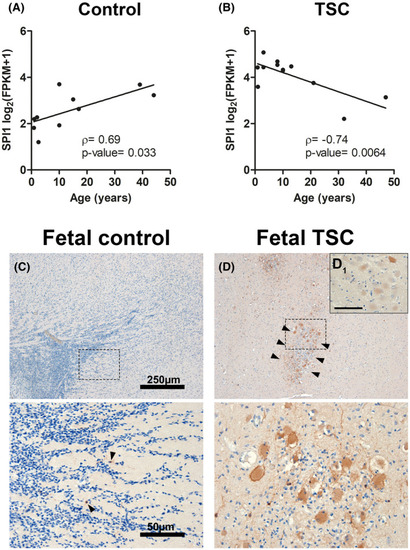
SPI1/PU.1 expression in malformed cells occurs early in TSC development. |
|
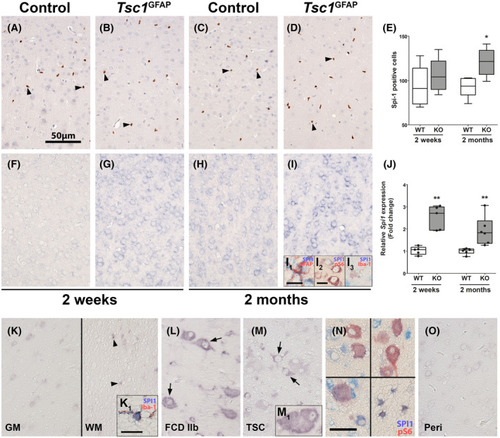
Spi1/Pu.1 RNA expression in the cortex is elevated in |
|
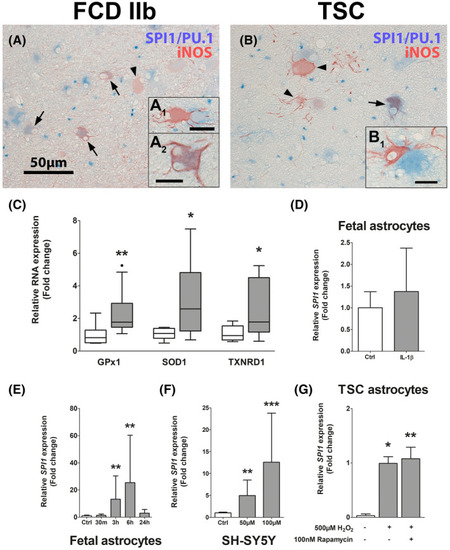
Oxidative stress is present in malformed cells in TSC cortical tubers and stimulates |