- Title
-
A Bioassay-Based Approach for the Batch-To-Batch Consistency Evaluation of Xuesaitong Injection on a Zebrafish Thrombosis Model
- Authors
- Ma, X., Chen, Y., Jiang, S., Zhao, X.
- Source
- Full text @ Front Pharmacol
|
|
|
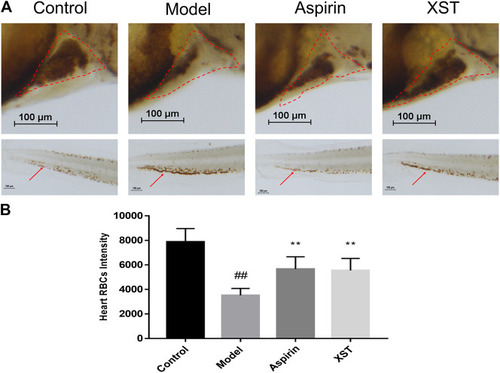
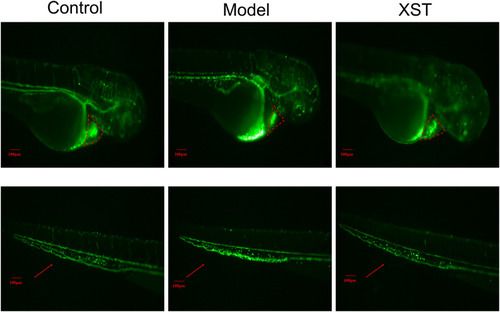
Transgenic LCR-GFP zebrafish in the control group, model group (AA 40 µmol/L), and XST group (AA 40 µmol/L + XST 400 µg/ml), RBCs in heart were marked by red dashed lines, caudal venous thrombus were guided by red arrows. n = 10. |
|
|
|
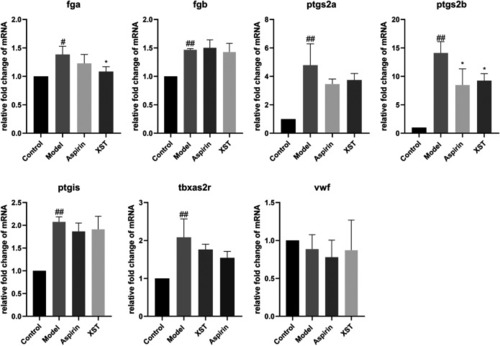
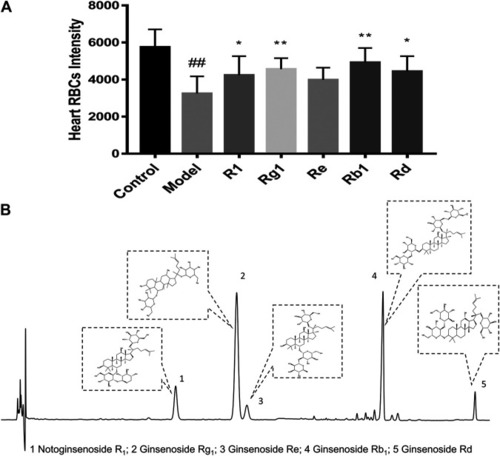
Relative fold change of mRNA of fga, fgb, ptgs2a, ptgs2b, ptgis, tbxa2r, and vwf in the control group, model group (AA 40 µmol/L), aspirin group (AA 40 µmol/L + aspirin 22.5 µg/ml), and XST group (AA 40 µmol/L + XST 400 µg/ml), n = 20. All data were expressed by the mean ± SD, ##p < 0.01, #p < 0.05 vs. control group; *p < 0.05 vs. model group. |
|
|
|
|
|
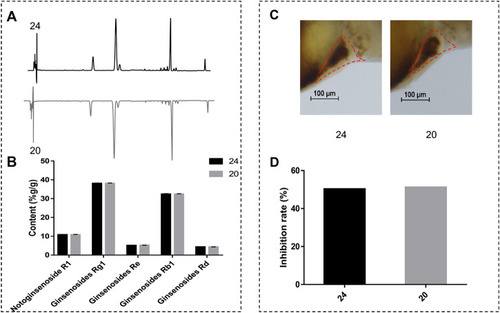
(A) The distinction between area normalization percent content and inhibition rates of 24 normal batches of XST and five abnormal batches of XST. (B) The distinction between the inhibition rates of 24 normal batches of XST and five abnormal batches of XST. |