- Title
-
Torpedo californica acetylcholinesterase is stabilized by binding of a divalent metal ion to a novel and versatile 4D motif
- Authors
- Silman, I., Shnyrov, V.L., Ashani, Y., Roth, E., Nicolas, A., Sussman, J.L., Weiner, L.
- Source
- Full text @ Protein Sci.
|
Thermal inactivation of |
|
Effect of |
|
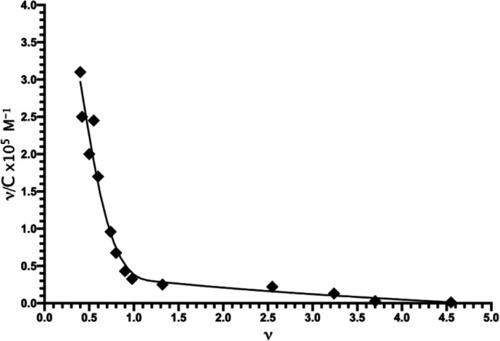
Scatchard plot of the interaction of Mn+2 with |
|
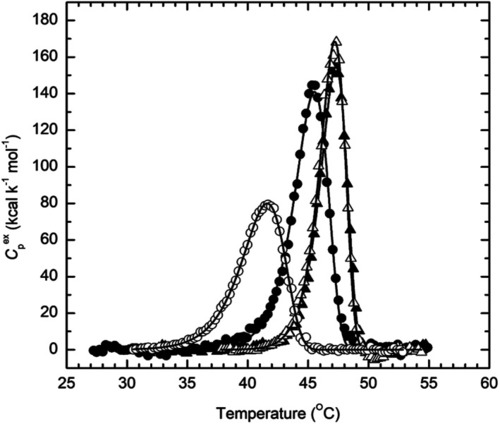
Temperature dependence of the excess molar heat capacity of |
|
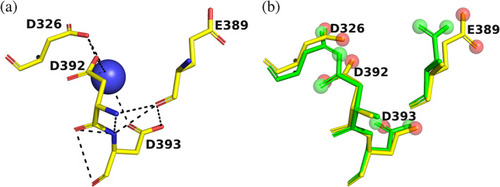
4D motif in |
|
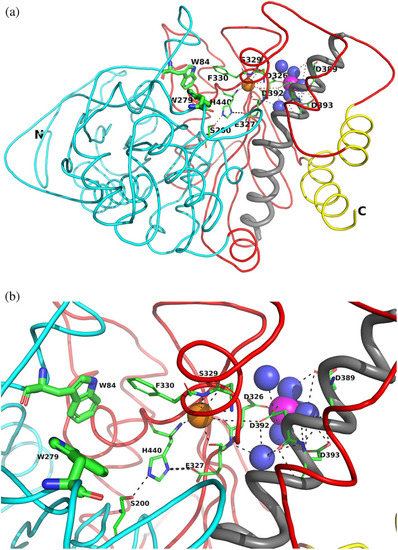
Overall views of the Mg+2/TcAChE complex. (a) Ribbon diagram of the Mg+2/TcAChE complex. The representation shows the entire structure, with the first subdomain, residues 4–305, in cyan, and the second, residues 306–535, in red. It is oriented looking into the active‐site gorge, with W279, in the peripheral anionic site (PAS), at the top of the gorge, and W84, in the catalytic anionic site (CAS) toward the back, adjacent to the catalytic triad, S200‐E327‐H440. All these residues are depicted as sticks. The long α‐helix, N383‐K413, against which the 4D pocket is glued, is in grey, and the two helices that contribute to the four‐helix bundle of the dimer, D365‐Y375 and V518‐T535, are in yellow. The Mg+2 in the 4D pocket is in magenta, and is surrounded by four waters in blue. A conserved water H‐bonds with D326, of the 4D motif, with E327 and H440, in the catalytic triad, and with the main‐chain nitrogen of F330, which, in turn, contributes to the CAS. This water which is homologous to water 623 in Koellner et al.,29 is shown as an orange sphere. (b) Close up, with the same orientation, showing the interactions of the active site, the 4D pocket, and the conserved water, shown as an orange sphere |
|
Sequence alignments of residues 320–400 in several AChEs and in HuBChE. The numbering used is that of TcAChE. Fully conserved residues are in white on a red background. The columns for the four residues corresponding to the 4D motif in TcAChE and zebrafish AChE are framed in green, and it can be seen that the motif is conserved only in these three AChEs. TcAChE, Torpedo californica AChE; TmAChE, Torpedo marmorata AChE; EeAChE, Electrophorus electricus AChE; DrAChE, Danio rerio AChE; BfAChE, Bungarus fasciatus AChE; HuAChE, human AChE; BoAChE, bovine AChE; MoAChE, mouse AChE; HdAChE, designed HuAChE, D4 variant38; HuBChE, human BChE; AgAChE, Anopheles gambiae AChE; DmAChE, Drosophila melanogaster AChE |
|
Pocket in BfAChE that is homologous to the 4D pocket in TcAChE. (a) Crystal structure43 (PDB‐ID 4qww), showing three Asp residues, a Glu residue, and a water. (b) Overlay of the BfAChE pocket (yellow sticks) on the TcAChE pocket (green sticks) with the distal oxygens displayed as red and green balls, respectively |
|
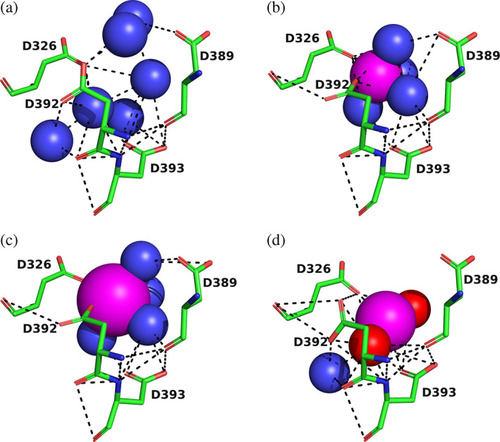
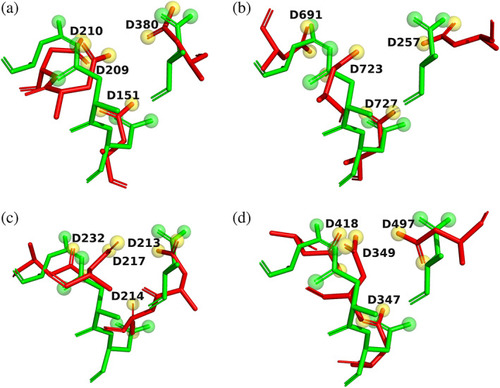
4D motifs in four proteins retrieved from the ASSAM server. The four Asp residues are shown as sticks, with carbons in green, oxygens in red, and nitrogens in blue. Solvent waters are shown as blue spheres, and the metal ions as magenta spheres, with their sizes proportionate to their Van der Waals radii. Noncovalent H‐bonds and ionic bonds are displayed as dashed black lines. (a) |
|
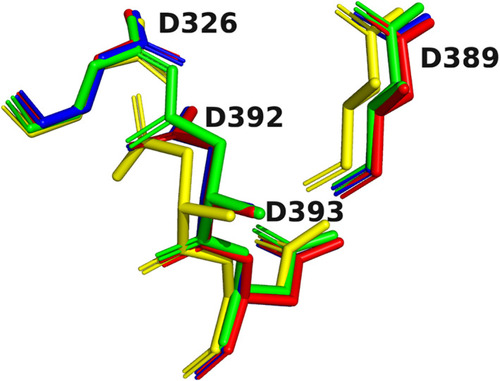
Overlays of the 4D motifs displayed in Figure |
|
Overlays on Apo |