- Title
-
PAK4 methylation by the methyltransferase SETD6 attenuates cell adhesion
- Authors
- Vershinin, Z., Feldman, M., Levy, D.
- Source
- Full text @ Sci. Rep.
|
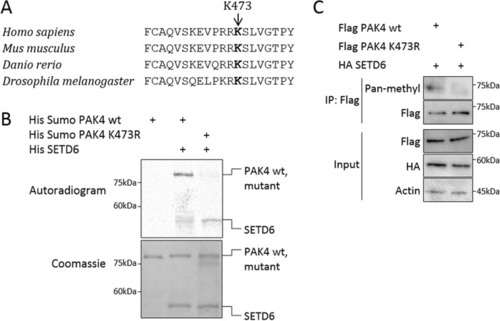
SETD6 methylates PAK4 at lysine 473. ( |
|
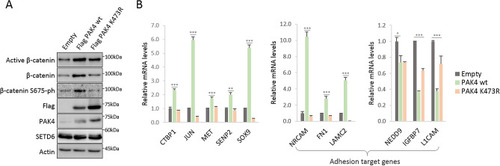
Methylated PAK4 at lysine 473 upregulates Wnt/β-catenin target genes. ( |
|
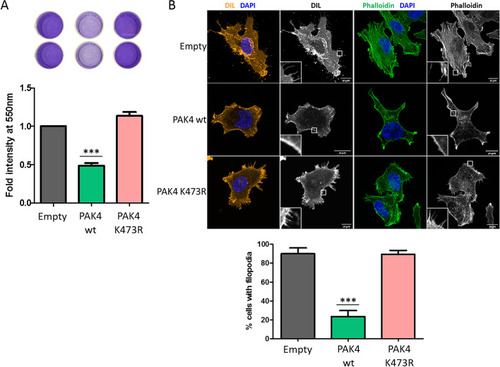
PAK4 K473me decreases adhesion of the cells and modulates cell morphology. ( |
|
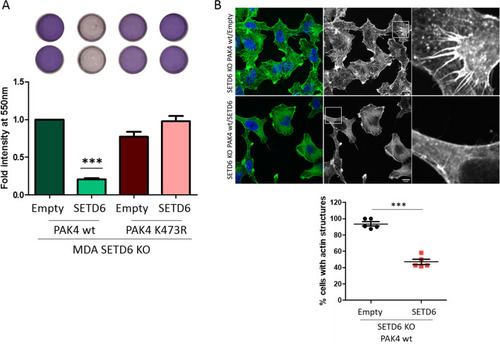
Cell adhesion properties are SETD6 dependent. ( |
|
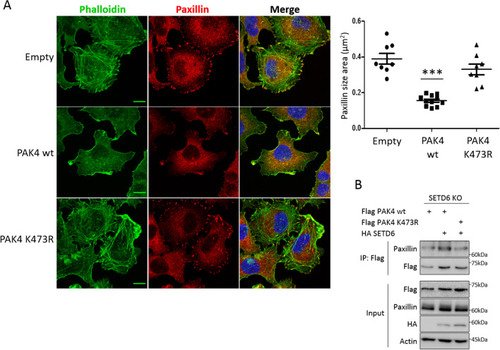
PAK4 K473me attenuates paxillin localization to focal adhesions. ( |
|
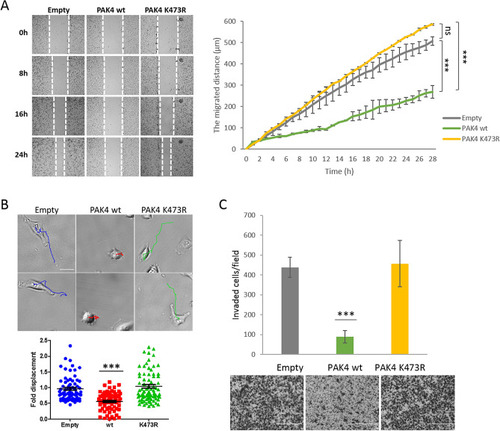
Unmethylated PAK4 promotes cell migration and cell invasion. ( |