- Title
-
Runx1 regulates zebrafish neutrophil maturation via synergistic interaction with c-Myb
- Authors
- Huang, Z., Chen, K., Chi, Y., Jin, H., Li, L., Zhang, W., Xu, J., Zhang, Y.
- Source
- Full text @ J. Biol. Chem.
|
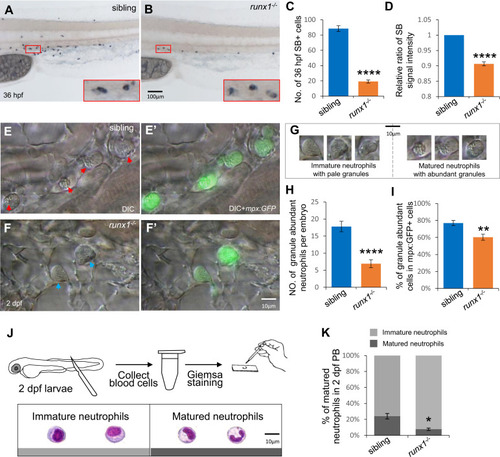
Neutrophil maturation was affected by runx1w84x mutation. A–D, SB staining showed decrease number and intensity of SB+ cells in 36-hpf runx1−/− mutants (B) compared with siblings. Red boxes show enlarged detail of SB+ cells in each group (×4). C, quantification of numbers of 36-hpf SB+ cells (each n ≥ 22). Neutrophils were counted based on the SB signals of the whole embryos. D, relative ratio of SB signal intensity was calculated in runx1 mutants and siblings. E–H, granule status in mpx:GFP+ neutrophils. (E, F, E’ and F’) In vivo VE DIC microscopy revealed reduction of granules in neutrophils in 2-dpf runx1 mutants (F and F’) compared with siblings (E and E’) in Tg(mpx:GFP) background (each n ≥ 22). Left panels are bright field DIC images. Right panels are overlays of bright field DIC images with corresponding GFP fluorescent images. Red arrowheads indicate matured neutrophils. Blue arrowheads indicate immature neutrophils. G, representative images of matured neutrophils with abundant granules and immature neutrophils with pale granules. H and I, quantification of absolute numbers of granule-abundant neutrophils per embryo (H) and relative percentage of granule-abundant cells in total mpx:GFP+ neutrophils (I). J and K, May–Grünwald–Giemsa staining of neutrophils in 2-dpf embryos (J) and neutrophils were quantitated by morphology (K). Gray and dark gray represent immature and mature neutrophils by their nucleic morphology. Scale bars, 100 μm (A and B), and 10 μm (E, F, E’, F’, G, and J). |
|
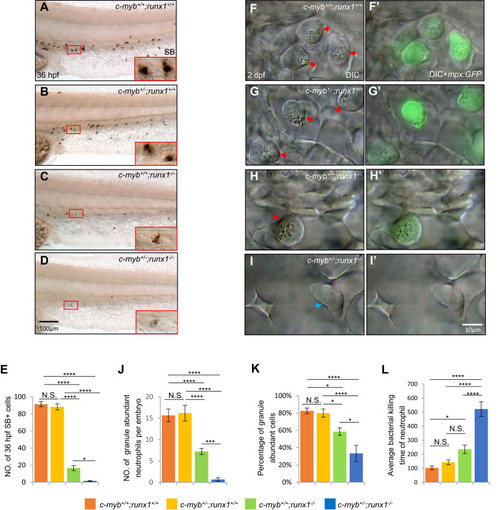
c-Myb and Runx1 synergistically regulate neutrophil maturation. A–E, SB staining showed further decrease in intensity and number of SB+ cells in 36-hpf c-myb+/−;runx1−/− double mutants (D) compared with single mutants. Red boxes show enlarged detail of SB+ cells in each group (×4). E, quantification of numbers of 36-hpf SB+ cells (each n ≥ 18). (F–I and F’–I’) In vivo VE DIC microscopy revealed further reduction of granules in neutrophils in 2-dpf double mutants (I and I’) compared with single mutants (G, H, G’ and H’) in Tg(mpx:GFP) background (each n ≥ 15). Red arrowheads indicate matured neutrophils with abundant granules. Blue arrowheads indicate immature neutrophils with pale granules. J–K, quantification of absolute numbers of granule abundant-neutrophils per embryo (J) and relative percentage of granule-abundant cells in total mpx:GFP+ neutrophils (K), each n ≥ 15. Scale bars equal 10 μm. L, neutrophil bacterial clearance time in each group. Scale bars, 100 μm (A–D), and 10 μm (F–I and F’–I’). |
|
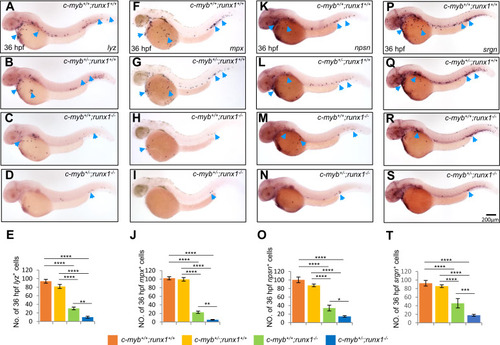
Genetic interaction between c-myb and runx1 on neutrophil-specific genes. A–D, WISH showed further decrease of lyz expression in 36-hpf double mutants (D) compared with single mutants (B and C). E, quantification of numbers of 36-hpf lyz+ cells (each n ≥ 16). F–J, WISH showed further decrease of mpx+ cells in 36-hpf double mutants (I) compared with single mutants. J, quantification of numbers of 36-hpf mpx+ cells (each n ≥ 20). K–N, WISH showed further decrease of npsn+ cells in 36-hpf double mutants (N) compared with single mutants. O, quantification of numbers of 36-hpf npsn+ cells (each n ≥ 8). P–S, WISH showed further decrease of srgn+ cells in 36-hpf double mutants (S) compared with single mutants. T, quantification of numbers of 36-hpf srgn+ cells (each n ≥ 8). Blue arrowheads indicate lyz+, mpx+, srgn+, and npsn+ neutrophils in each row. Scale bars, 200 μm. |
|
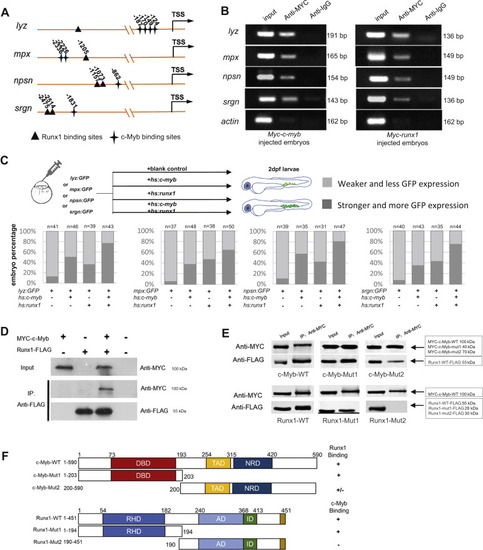
c-Myb and Runx1 synergistically regulate neutrophil-specific genes transcription. A, schematic diagram of the lyz, mpx, srgn, and npsn promoter region. The transcription starting site is designated as TSS. Putative c-Myb and Runx1 binding sites are marked by stars and triangles respectively using JASPAR online software. B, ChIP shows that MYC-tagged c-Myb and MYC-tagged Runx1 bind to the promoter region of the lyz, mpx, npsn, and srgn promoters. Lysates from the embryos injected with the Myc-c-myb (left panel) and Myc-runx1 plasmids (right panel) were precipitated with anti-MYC antibodies. The precipitates were then subjected to semiquantitative PCR analysis compared with anti-IgG control. Input DNA control was on the left of each panel. C, Upper panel shows representative procedures of GFP reporter assay. lyz(-2.4k):GFP, mpx(-8k):GFP, npsn(-2k):GFP and srgn(-5k):GFP coinjected with or without hs:c-myb, hs:runx1 plasmid. Two groups were classified by GFP fluorescence intensity and GFP+ cells number. Lower panel shows embryo percentage of each group (each n ≥ 20). Gray represents weaker GFP expression and less GFP+ cells. Dark gray represents stronger GFP expression and more GFP+ cells. D, in vitro coimmunoprecipitation experiment detected the interaction between c-Myb and Runx1. Myc-tagged c-myb and Flag-tagged runx1 were transfected into 293T cells as indicated and cell lysates were immunoprecipitated with anti-FLAG antibody. The immunoprecipitants were examined by western blot using anti-MYC and anti-FLAG antibodies. Input represents 10% of total cell lysates used for immunoprecipitation. E, deletions impinging on the c-Myb DBD (top panel) and Runx1 RHD (low panel) decrease the interaction between c-Myb and Runx1. MYC-c-Myb was immunoprecipitated, and western blots were probed with antibodies to MYC or FLAG. F, summary of c-Myb and Runx1 mapping experiments. AD, activation domain; DBD, DNA-binding domain; ID, inhibitory domain; NRD, negative-regulatory domain; RHD, RUNT-homology domain; TAD, transactivation domain. |
|
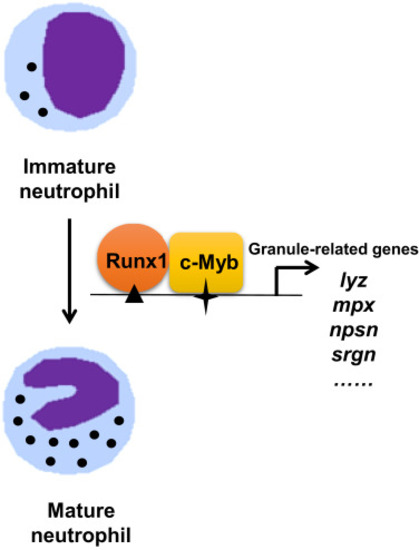
Models of neutrophil regulation by c-Myb and Runx1 of in zebrafish. Synergistic interaction of c-Myb and Runx1 in neutrophil maturation. c-Myb and Runx1 are essential for granule-related genes expression in neutrophil maturation. c-Myb and Runx1 bind to the neutrophil-specific genes promoter and interact to cooperatively regulate their expressions. |