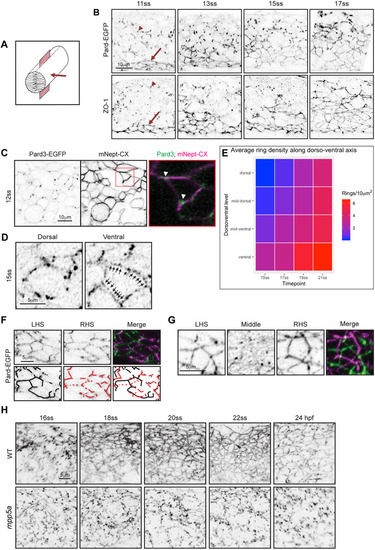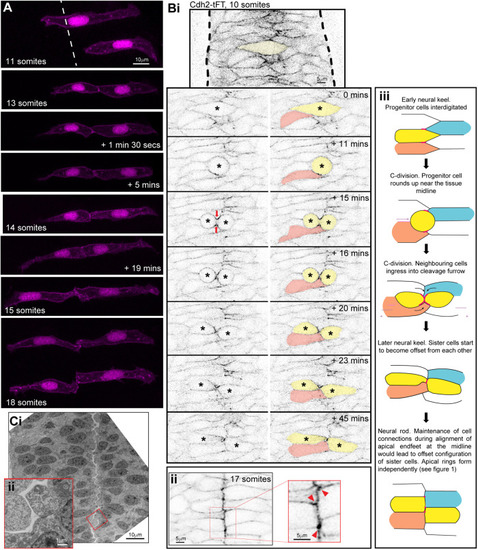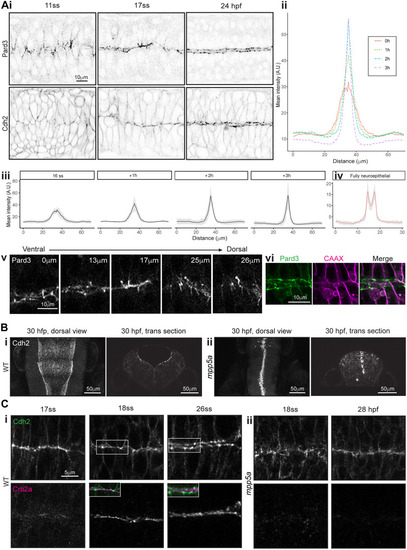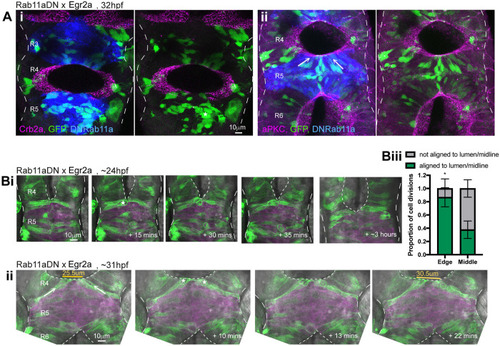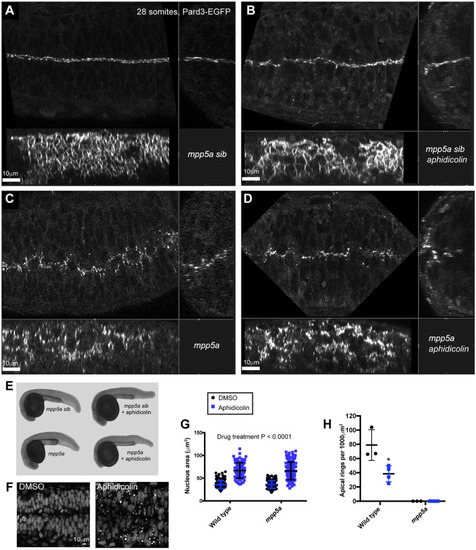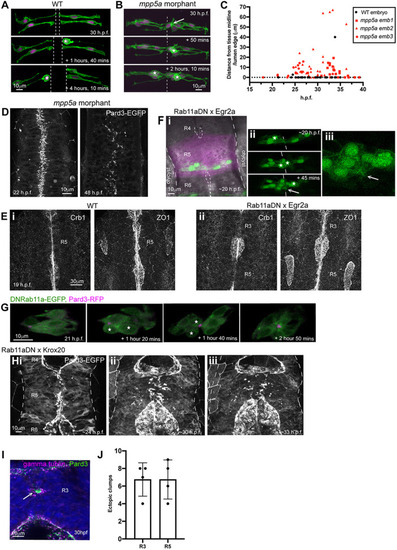
Persistent adhesion in Mpp5a- and Rab11a-deficient cells. (A) Images from time-lapse movie of mosaically labelled neuroepithelial cells in the hindbrain of a 30 hpf wild-type (WT) embryo, dorsal view. Cells have separated across the tissue midline and the lumen has opened (dashed lines). D-divisions (stars) occur at the apical surface, and cells re-established an elongated morphology towards the basal surface after division. (B) Images from time-lapse movie of mosaically labelled neuroepithelial cells in the hindbrain of a 30 hpf mpp5a morphant embryo, dorsal view. Cells have failed to separate across the tissue midline (dashed line) and have formed clumps (sometimes with a rosette-like structure; arrow). D-divisions (stars) occurred near the centre of the cell clumps, which was often not situated near the tissue midline. Of 117 daughter cells followed post-division, ≥40% did not re-extend fully to the basal side of the neural rod. (C) Graph of cell division locations in relationship to the tissue midline or lumen edge over development. Seventy-one cells were analysed from three mpp5a morphant embryos. In embryos >25 hpf, 49% of mpp5a morphant cells divided 5 µm or more away from the midline. Division locations from a single wild-type embryo example are included in black. (D) A 10 µm z-projection, dorsal view, at mid-dorsoventral level through the hindbrain of mpp5a morphant Pard3-EGFP embryos at 22 and 48 hpf. At 22 hpf, Pard3-EGFP was localised near the tissue midline but did not form continuous straight expression domains as seen in wild types (Fig. 3Ai), and the lumen failed to open. By 48 hpf, Pard3-EGFP localisation became fragmented into clumps. Nine of nine mpp5a mutant embryos and six of eight mpp5a morphant embryos >24 hpf old from five different experiments had fragmented midlines and ectopic apical proteins. The extent of this disorganisation was greater in older embryos. Two of six mpp5a morphant embryos had a milder phenotype (see Fig. S1). Nine of nine wild-type embryos had normal apical surfaces, with no ectopic apical proteins. (E) 70-80 µm maximum projections, dorsal view of hindbrain at 19 hpf, stained for Crb1 and ZO-1. (Ei) Wild-type embryo. Both Crb1 and ZO-1 were localised to the apical midline (n=3/3). (Eii) Embryo in which UAS:DNRab11a was expressed under the Egr2a:KalTA4 activator in rhombomeres (R) 3 and 5. ZO-1 localised to the midline, but Crb1 was largely absent from rhombomeres 3 and 5 (n=3/3). (Fi) 11 µm maximum-intensity projection of dorsal view of hindbrain neuroepithelial cells at 20 hpf in a DNRab11a × Egr2a embryo. Magenta shows DNRab11a rhombomere 5. Lumen in rhombomere 5 failed to open but had opened in rhombomeres 4 and 6 (short-dashed lines are lumen surface, long-dashed lines are basal surface). (Fii) Maximum-intensity projection images from time-lapse movie of cells in Fi, which failed to separate across the tissue midline (white dashes). Similar to mpp5a morphant embryos (B), after D-divisions (stars), cells remained attached and were arranged into a rosette-like structure (arrow), enlarged in Fiii. Cell clumping or rosette-like structures were observed in DNRab11a rhombomeres of all nine embryos analysed from three experiments. Note, in all panels in F we have edited out overlying cells from z-planes that would otherwise obscure the view of the cells participating in the rosette. Unedited versions of these images can be seen in Fig. S1. (G) Time-lapse reconstruction showing mosaically distributed DNRab11a-EGFP neuroepithelial cells in hindbrain of 24-somite-stage (21 hpf) embryo. After D-divisions (stars), cells did not separate and formed a rosette around centrally located puncta of Pard3-RFP. (H) 70 µm maximum projections from time-lapse movie through a DNRab11a × Egr2a embryo hindbrain labelled with Pard3-EGFP, starting at 24 hpf. As in mpp5a morphants in D, Pard3 was initially close to the midline of rhombomere 5, but the lumen failed to open and Pard3 localisation became fragmented into clumps that dispersed over time (17 of 17 embryos). (I) 4 µm maximum projection, dorsal view of 30 hpf DNRab11a × Egr2a embryo rhombomere 3, stained for Pard3 and gamma-tubulin. Pard3 is localised in a round clump, surrounded on all sides by centrosomes. (J) DNRab11a rhombomeres 3 and 5 had approximately seven ectopic clumps of Pard3 and gamma-tubulin at 30 hpf (n=4 embryos). Error bars denote standard deviations. No ectopic clumps were seen in wild-type rhombomeres.
|