|
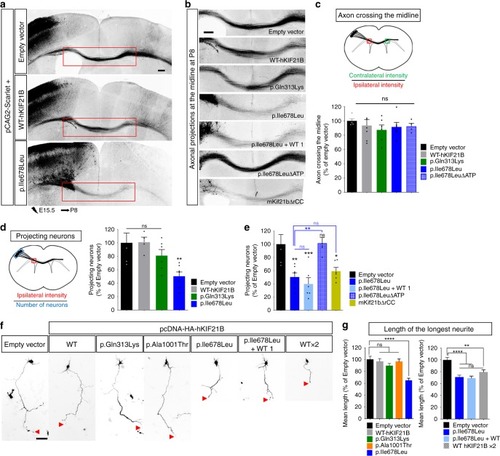
p.Ile678Leu hKIF21B variant impedes inter-hemispheric connectivity.a Coronal sections of P8 brains after IUE with pCAG2-Scarlet and indicated NeuroD-IRES-GFP constructs. b Close-up views of the red boxed area in a showing impaired axonal inter-hemispheric connectivity upon expression of the p.Ile678Leu variant or the hyperactive mKif21bΔrCC but not upon expression of WT, p.Gln313Lys or immotile p.Ile678LeuΔATP. Rescue experiments were done by co-expressing p.Ile678Leu-hKIF21B (1 µg/µL) together with WT-hKIF21B constructs at 1 µg/µL (p.Ile678Leu + WT 1). Scale bars, 250 μm. c, d Upper (c) or left (d) panels, schematic describing methods used to quantify the percentage (c) of axon crossing the midline and (d) of projecting neurons. Lower (c) or right (d) panels, histograms presenting the percentage (c) of axon crossing the midline and (d, e) of projecting neurons. Data (means ± S.E.M.) were analyzed by one-way ANOVA (Bonferroni’s multiple comparisons test). Number of pups analyzed: c Empty vector, n = 5; WT, n = 7; p.Gln313Lys, n = 6; p.Ile678Leu, n = 6; p.Ile678LeuΔATP, n = 4; d, e empty vector, n = 5; WT, n = 4; p.Gln313Lys, n = 6; p.Ile678Leu, n = 7; p.Ile678LeuΔATP, n = 5; p.Ile678Leu + WT, n = 6; mKif21bΔrCC, n = 6. f Representative DIV2 cortical neurons transfected at DIV0 with pCAG2-Scarlet together with empty pcDNA-HA or WT (at 1 (WT) or 2 µg/µL (WT ×2)) or mutant pcDNA-HA-hKIF21B constructs. Rescue experiments were done by co-expressing mutated p.Ile678Leu hKIF21B variant together with NeuroD-WT-hKIF21B (ratio 1:1; p.Ile678Leu + WT 1). Red arrowheads point to the axon tip. g Quantification of the longest neurite length (axon) at DIV2. Bars represent the means of the longest neurite length ± S.E.M. Significance was calculated by one-way ANOVA (Bonferroni’s multiple comparisons test). Number of neurons analyzed: (left graph) Empty vector, n = 94; WT, n = 96; p.Gln313Lys, n = 104; p.Ile678Leu, n = 144; p.Ala1001Thr, n = 133, from four (Empty vector, WT, p.Ile678Leu) or three (p.Gln313Lys, p.Ala1001Thr) independent experiments; (right graph) empty vector, n = 132; WT 2 µg/µL, n = 125; p.Ile678Leu + WT, n = 146; p.Ile678Leu, n = 134; from four independent experiments. ns non-significant; *P < 0.05; **P < 0.005; ***P < 0.001; ****P < 0.0001. Scale bars, (a) 200 µm, (b) 250 µm (f) 50 µm. Source data are provided in the Source Data file.
|