- Title
-
SPLICS: a split green fluorescent protein-based contact site sensor for narrow and wide heterotypic organelle juxtaposition
- Authors
- Cieri, D., Vicario, M., Giacomello, M., Vallese, F., Filadi, R., Wagner, T., Pozzan, T., Pizzo, P., Scorrano, L., Brini, M., Calì, T.
- Source
- Full text @ Cell Death Differ.
|
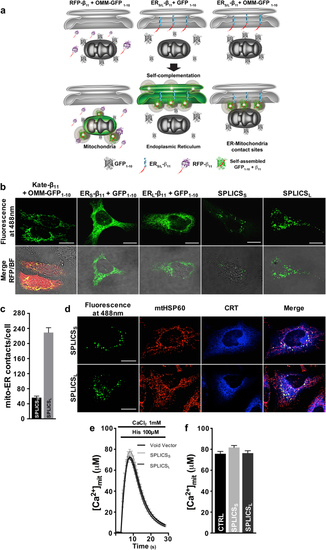
Functional characterization of the SPLICS probes. |
|
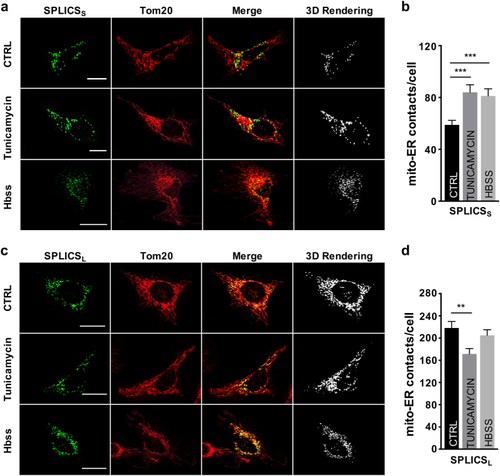
Effect of Tunicamycin and Hbss treatment on ER–mitochondria contacts. Immunofluorescence against mitochondria (Tom20, red) is shown in the panels on the middle. The green channel is the merge of several planes. Scale bar 20 µm. |
|
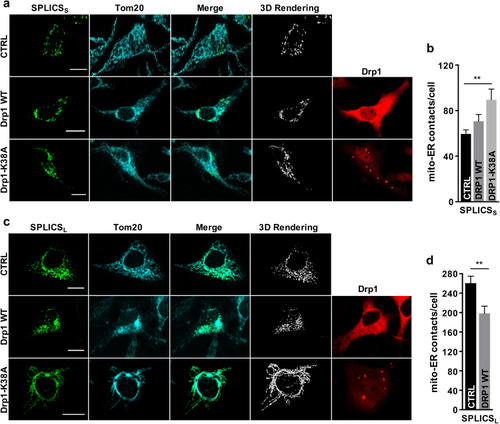
Effects of Drp1 overexpression on ER–mitochondria contacts. Immunofluorescence against mitochondria (Tom20, cyan) and Drp1 (red) is shown in the corresponding panels. The green channel is the merge of several planes. Scale bar 20 µm. |
|
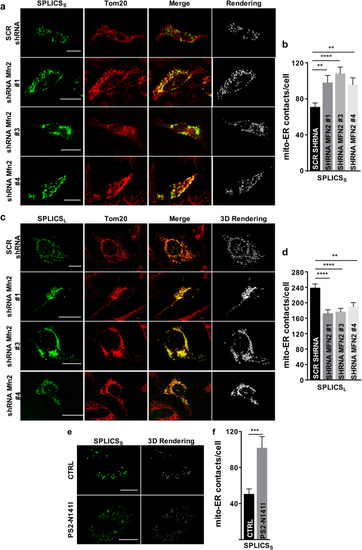
Effect of Mfn2 knockdown and mutant PS2 on ER–mitochondria interface. Immunofluorescence against mitochondria (Tom20, red) is shown in the panels on the middle. The green channel is the merge of several planes. Scale bar 20 µm. |
|
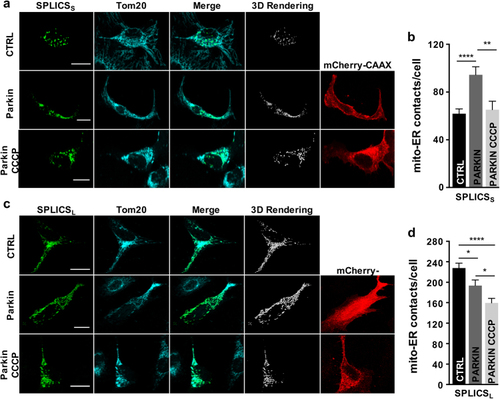
Effects of Parkin on ER–mitochondria contacts. Immunofluorescence against mitochondria (Tom20, cyan) is shown in the panels on the middle. The green channel is the merge of several planes. Scale bar 20 µm. |
|
Expression of the SPLICSS probe in living zebrafish embryos. |