- Title
-
GIPC proteins negatively modulate Plexind1 signaling during vascular development
- Authors
- Carretero-Ortega, J., Chhangawala, Z., Hunt, S., Narvaez, C., Menéndez-González, J., Gay, C.M., Zygmunt, T., Li, X., Torres-Vázquez, J.
- Source
- Full text @ Elife

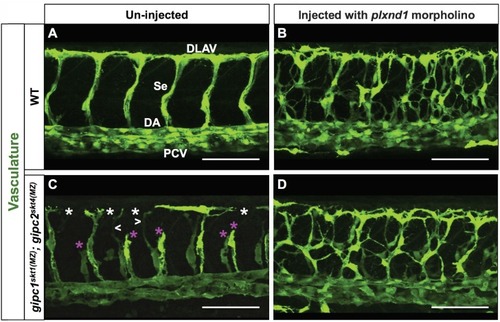
ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. |

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. PHENOTYPE:
|

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. |

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. |
|
( |
|
( |
|
( |
|
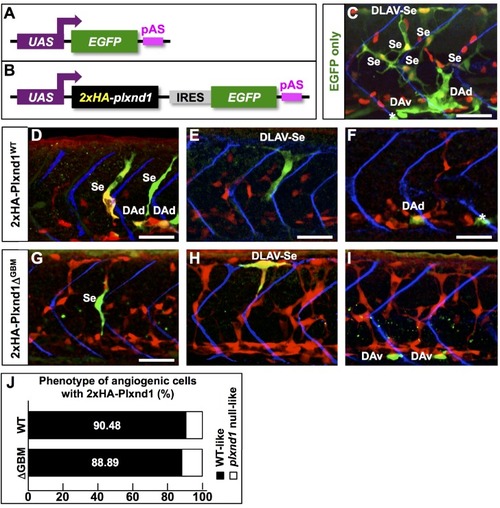
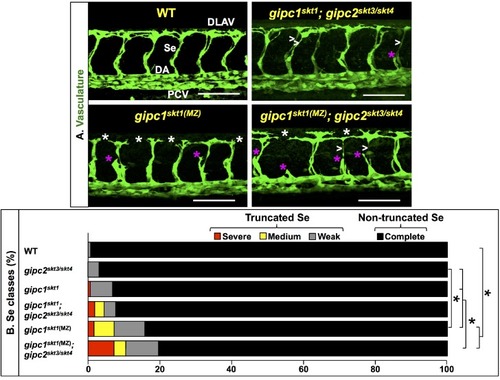
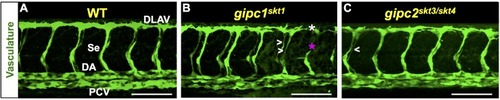
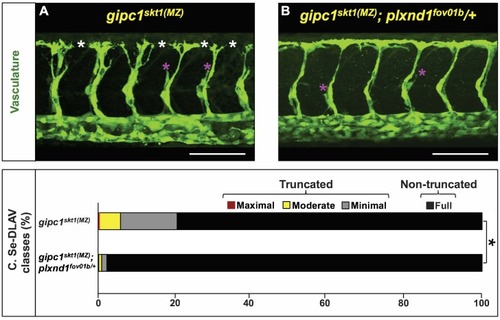
In the mutant proteins, the red bar denotes novel amino acid sequences resulting from mutagenic frameshift(s). See |
|
( |
|
( |
|
( PHENOTYPE:
|