- Title
-
Autoregulatory loop and retinoic acid repression regulate pou2/pou5f1 gene expression in the zebrafish embryonic brain
- Authors
- Parvin, M.S., Okuyama, N., Inoue, F., Islam, M.E., Kawakami, A., Takeda, H., and Yamasu, K.
- Source
- Full text @ Dev. Dyn.
|
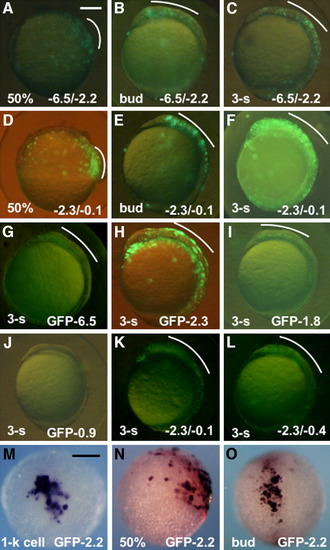
Regulatory activities of the subregions in the upstream DNA of pou2. A-F: Expression of the reporter (GFP-0.1) under regulation of the -6.5/-2.2 (A-C) and -2.3/-0.1 (D-F) fragments at 50% epiboly (A,D), bud stage (B,E), and three-somite stage (C,F). G-J: Expression of the green fluorescent protein (GFP) constructs under regulation by different regions of the pou2 upstream DNA at early somite stages. K,L: Expression of GFP-0.1 co-injected with -2.3/-0.1 (K) and -2.3/-0.4 (L) at early somite stages. M-O: mRNA expression of GFP-2.2, as revealed by whole-mount in situ hybridization, at the 1-k cell (M), 50% epiboly (N), and bud (O) stages. GFP expression in the dorsal blastoderm and head regions are marked with white curves. A-L,N: Lateral views with anterior to the top and dorsal to the right. M: Animal view. O: Dorsal view with anterior to the top. Scale bar = 200 μm. |
|
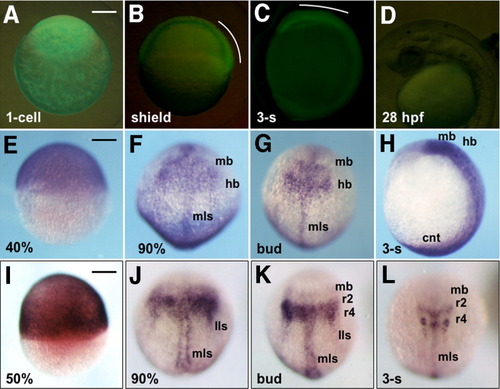
Stable expression of GFP-6.5 in transgenic embryos. A-C: Expression of GFP-6.5 as visualized with green fluorescent protein (GFP) fluorescence in a fertilized egg (A), or in developing embryos at the shield (B) and three-somite (C) stages. D: GFP expression could not be detected at 28 hours postfertilization (hpf) in the head. GFP expression in the dorsal blastoderm and head regions is marked with white curves. E-H: Expression of GFP-6.5 mRNA, detected by whole-mount in situ hybridization, in embryos at 40% epiboly (E), 90% epiboly (F), bud (G), and three-somite (H) stages. I-L: Endogenous expression of pou2 at 50% epiboly (I), 90% epiboly (J), bud (K), and three-somite stages (L). A,E,I: Lateral views with animal poles to the top. B-D,H: Lateral views with anterior to the top and dorsal to the right (B,C,H) or with anterior to the left and dorsal to the top (D). F,G,J-L: Dorsal views with anterior to the top. cnt, caudal neural tube; hb, hindbrain; lls, lateral longitudinal stripe; mb, midbrain; mls, medial longitudinal stripe. Scale bar = 200 μm. |
|
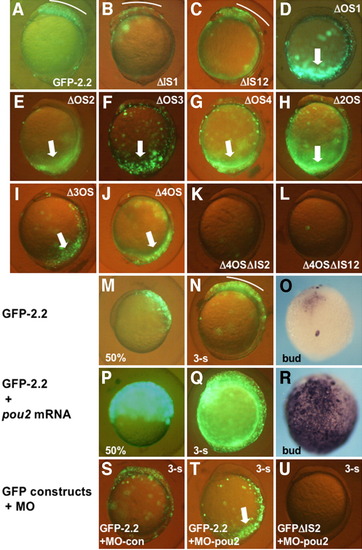
Roles of the octamer sequences in the transcriptional regulation by the proximal upstream DNA. A-L: GFP-2.2 or its mutated constructs were introduced into embryos and examined for their green fluorescent protein (GFP) expression at early somite stages. Deletion of the intervening sequences little affected the expression (B,C), while disruption of any of the four octamer sequences abrogated the brain expression, but gave rise to ectopic expression in the caudal region (thick arrows, D-J). Deletion of IS2 from GFP-2.2 lacking the octamer sequences eliminated the ectopic expression in the caudal region (K,L). M-R: The expression of GFP-2.2, which was restricted to the dorsal blastoderm at 50% epiboly (M), and to the brain at the bud (O) and three-somite stages (N), was expanded significantly by pou2 overexpression by mRNA injection (P-R), as was revealed both by GFP fluorescence (M,N,P,Q) and whole-mount in situ hybridization (O,R). S-U: The expression of GFP-2.2 (S,T) and GFPΔIS2 (U) in embryos injected with MO-con (S) or MO-pou2 (T,U). |
|
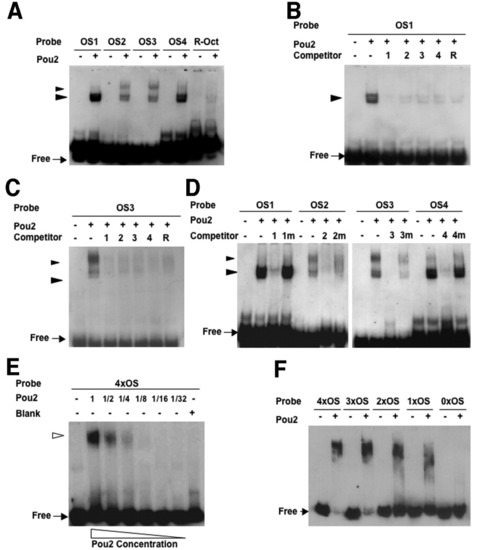
Binding activities of the octamer sequences in the pou2 regulatory region with the Pou2 protein. A: Digoxigenin (DIG) -labeled probes for the four octamer sequences (OS1-4) and R-Oct sequence generated shifted bands in the presence of Pou2. The fast-migrating doublet bands marked with large arrowheads were generated by all the probes, whereas the slower-migrating bands, shown with small arrowheads, were seen only when the pou2 octamer sequences were used. B,C: Binding of Pou2 with DIG-labeled OS1 (B) and OS3 (C) was competed out with 100-fold molar excess of the unlabeled oligos (OS1-4 and R-Oct). D: Binding of Pou2 with the four octamer probes was competed with 100-fold excess of cognate oligos (OS1-4), but not at all or only partially by mutated oligos (OS1m-4m). E: Binding of Pou2 with 4×OS DNA containing the four octamer sequences (OS1-4) generated a single shifted band with no intermediate bands. F: Binding of Pou2 with 4xOS, 3xOS, 2xOS, 1xOS, and 0xOS, which included decreasing numbers of the OS sequences, was examined, showing a gradual decrease in size of the complex and an increase in the amount of free probes. |
|
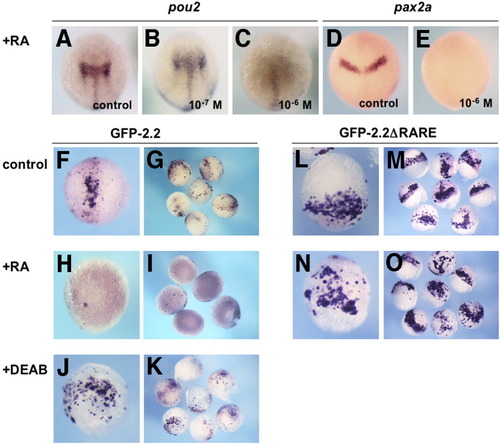
Repression of pou2 and GFP-2.2 expression by retinoic acid (RA). The mRNA expression of pou2 and pax2a in uninjected embryos (A-E) and green fluorescent protein (GFP) constructs in injected embryos (F-O) was examined by whole-mount in situ hybridization. A-C: Expression of endogenous pou2 mRNA at the bud stage in the mid-hindbrain (A) was expanded by 10-7 M RA (B), whereas repressed effectively by 10-6 M RA (C). D,E: Expression of pax2a mRNA at the bud stage in the MHB region (D) was repressed by 10-6 M RA (E). F-I: Expression of GFP-2.2 in injected late gastrulae, which was seen in the mid-hindbrain region (F,G), was effectively repressed by 10-6 M RA (H,I). J,K: GFP-2.2 expression in injected embryos expanded laterally in the posterior region by diethylaminobenzaldehyde (DEAB) treatment. L,M: Expression of GFP-2.2ΔRARE in injected embryos was expanded laterally near the blastoderm margin compared with that of GFP-2.2. N,O: Expression of GFP-2.2 ΔRARE in embryos treated with 10-6 M of RA. The observed expression pattern was indistinguishable from that in untreated embryos (L,M). A-F,H,J,L,N: Dorsal views with anterior to the top. RARE, retinoic acid-responsive element. EXPRESSION / LABELING:
|
|
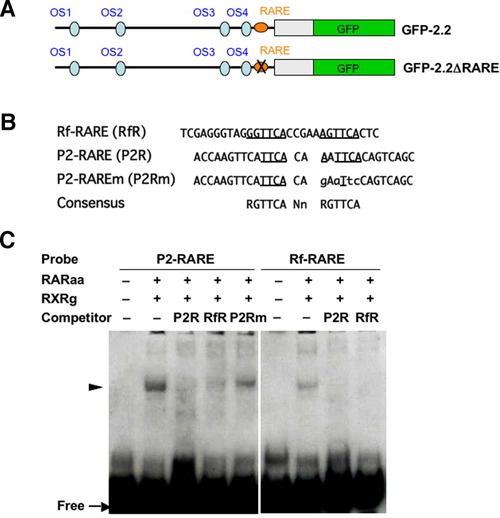
Binding of the DR2-type retinoic acid-responsive element (RARE) with the RAR/RXR complex. A: Schemes of the GFP-2.2 constructs that are intact or possesses a disrupted RARE (GFP-2.2ΔRARE). B: Sequences of the three oligos, Rf-RARE, P2-RARE, and P2-RAREm, are aligned. Sequences corresponding to the RARE consensus, which is shown at the bottom, are underlined. The bases substituted in P2-RAREm compared with the original oligo (P2-RARE) are shown with lower-case letters. C: Electrophoretic gel mobility shift assay (EMSA) showing specific binding of RAR/RXR with P2-RARE. Both P2-RARE and Rf-RARE formed complexes with RAR/RXR, which were competed out by a 100-fold molar excess of P2-RARE and/or Rf-RARE, but not by P2-RAREm. |