- Title
-
Cnbp ameliorates Treacher Collins Syndrome craniofacial anomalies through a pathway that involves redox-responsive genes
- Authors
- de Peralta, M.S., Mouguelar, V.S., Sdrigotti, M.A., Ishiy, F.A., Fanganiello, R.D., Passos-Bueno, M.R., Coux, G., Calcaterra, N.B.
- Source
- Full text @ Cell Death Dis.
|
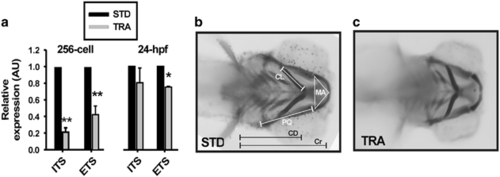
Tcof1 knockdown in zebrafish recapitulates TCS features. (a) RT-qPCR of rRNA transcripts (by ETS and ITS quantification) in samples from control (STD) and TRA-MO-injected 256 cell stage and 24 hpf embryos. Bars represent the mean of relative abundance and S.E.M., n=3, *P<0.05, **P<0.01 (t-test). AU: arbitrary units. (b and c) Ventral views of a typical control (b; STD-MO-injected) and a representative TCS-like (c; TRA-MO-injected) 4 dpf larvae stained with Alcian Blue. Meckel area (MA), area of the inner triangle defined by the Meckel cartilage, PQ length: length of palatoquadrate+hyosymplectic cartilages, ceratohyal length (CL): length of ceratohyal cartilage, CD: distance between ceratohyal cartilages joint and lateral fins, cranial distance (Cr): distance between the most anterior Meckel and lateral fins PHENOTYPE:
|
|
Craniofacial phenotypes generated by tcof1 knockdown were partially rescued by Tp53 depletion. (a) Western blotting showing the Tp53 abundance in control (STD), TRA and TRA+ATGTp53 24 hpf embryos. Molecular weight markers are shown at the right. Tubulin was analyzed as a loading control. The graph shows the densitometric quantification of western blotting band (normalization of Tp53 signal with respect to tubulin). Bars represent mean of relative abundance and S.E.M., n=3, *P<0.05 (t-test) AU: arbitrary units. (b) Expression analysis of Tp53 targets: ccng1 (encoding cyclin G1), pmaip1 (encoding Noxa) and tp53inp1 (encoding tumor protein p53-inducible nuclear protein 1) in control (STD), TRA and TRA+ATGTp53 24 hpf embryos. Bars represent mean of relative abundance and S.E.M., n=3, *P<0.05, (ANOVA test). (c) Acridine Orange staining in vivo performed in 24 hpf embryos. Representative photos of embryos displaying normal (+), intermediate (++) or extensive (+++) cell death. The bar graph shows the distribution of each phenotype among the groups (STD, TRA and TRA+ATGTp53 embryos). ey=eye, n=50 embryos in each of three independent experiments. ***P<0.001 versus STD; ###P<0.001 versus TRA; NS=non-significant P>0.05 (chi-square test). (d–h) Quantification of craniofacial parameters (as defined in Figure 1) measured in STD, TRA-MO TRA+ATGTp53 larvae (4 dpf), stained with Alcian Blue. (d) Meckel Area, (e) PQ, (f) Ceratohyal length, (g) CD, (h) Cr. Bars represent means in AU and S.E.M. n=47 for STD, n=79 for TRA, n=30 for TRA+ATGTp53. *P<0.05, **P<0.01, ***P<0.001 versus STD; #P<0.05, ##P<0,01 ###P<0.001 versus TRA; NS=non-significant P>0.05 (t-test) |
|
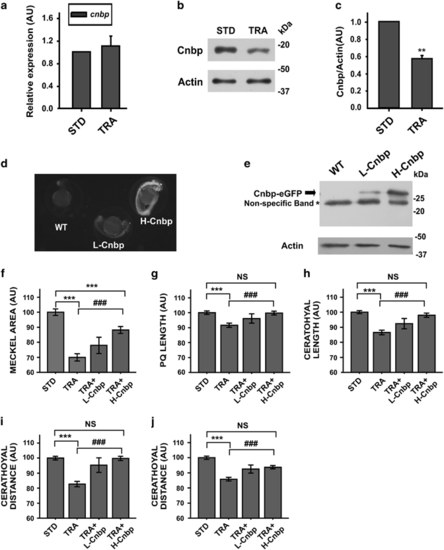
TCS-like phenotype is ameliorated in cnbp-overexpressing transgenic zebrafish. (a) Relative abundance of cnbp mRNA determined by RT-qPCR using total RNA from 24 hpf zebrafish control (STD) and TRA-embryos. Bars represent mean of relative abundances and S.E.M., n=3 AU: arbitrary units. (b) Western blotting analysing cnbp expression in 24 hpf embryos microinjected with TRA-MO. Controls were injected with STD-MO. Actin detection was used as a loading control. (c) Densitometric analysis of Cnbp normalized to Actin. The bars are means±S.E.M. of three experiments. n=5, **P<0.01 (t-test) (d) Fluorescence images of wild-type (WT) and transgenic zebrafish Tg(XIa.Eefiai:cnbpa-EGFP) lines (L-Cnbp and H-Cnbp). (e) Levels of Cnbp-eGFP detected by western blotting in 24 hpf transgenic zebrafish embryos by using anti-GFP antibodies. (*) Non-specific bands. Actin was assessed as a loading control. (f–j) Bar charts representing the quantification of craniofacial parameters measured in controls (STD), wild-type tcof1-knockdown larvae (TRA) and transgenic tcof1-knockdown larvae overexpressing low (TRA+L-Cnbp) or high (TRA+H-Cnbp) levels of Cnbp. (f) Meckel Area, (g) PQ length, (h) Ceratohyal length; (i) CD, (j) Cr. In TRA+L-Cnbp larvae, only the CD was restored (P<0.05 versus TRA and not different from STD), the Meckel area was indistinguishable from TRA and the rest of parameters showed in-between statistics (not displayed). Bars represent means in AU and S.E.M. n=47 for STD, n=79 for TRA, n=15 for TRA+L-Cnbp, n=36 for TRA+H-Cnbp. ***P<0.001 versus STD, ###P<0.001 versus TRA NS=non-significant P>0.05 (t-test) |