Fig 5
- ID
- ZDB-FIG-250722-29
- Publication
- Feltes et al., 2025 - Phenotype to genotype: A new and rapid approach using whole-genome sequencing
- Other Figures
- All Figure Page
- Back to All Figure Page
|
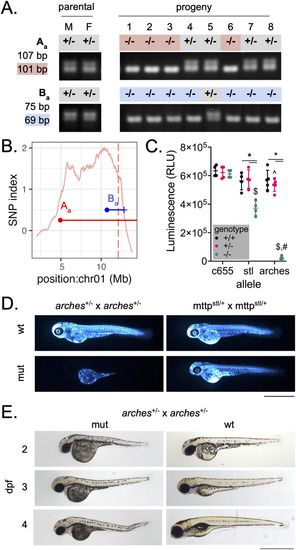
Recombinant mapping narrows region of interest to identify a loss-of-function allele of A) markers Aa and Ba were outputted by WheresWalker and used to genotype |