Figure 4
- ID
- ZDB-FIG-250615-14
- Publication
- Lin et al., 2025 - Phage-Antibiotic Synergy Enhances Biofilm Eradication and Survival in a Zebrafish Model of Pseudomonas aeruginosa Infection
- Other Figures
- All Figure Page
- Back to All Figure Page
|
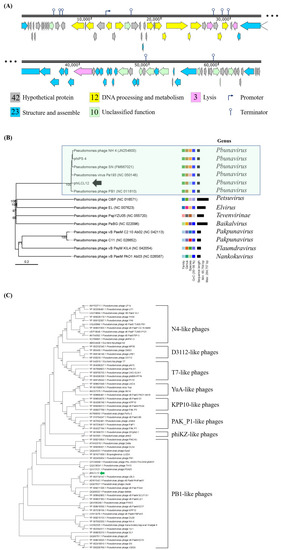
Genome map and phylogenetic analysis of phiLCL12 and related phages. ( |