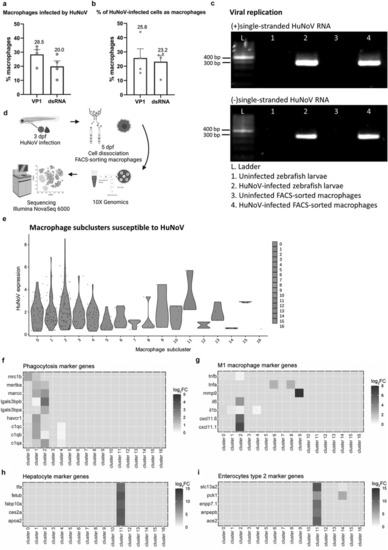
The in vivo tropism for macrophages is further confirmed by combined results of flow cytometry, RT-PCR on FACS-sorted macrophages, and a scRNA-seq analysis on dissected zebrafish larval intestines. Flow cytometry analysis on 2 dpi Tg(mpeg:mCherry-F) zebrafish larvae infected with HuNoV shows (a) the percentage of macrophages that contain HuNoV (either VP1 or dsRNA) and (b) the percentage of overall infected cells (VP1 or dsRNA) that are identified as macrophages. N = 4. Mean values ± SEM are shown. (c) Detection of (+)ss and (-)ss HuNoV RNA in both whole zebrafish larvae and FACS-sorted macrophages from HuNoV-infected Tg(mpeg:mCherry-F) zebrafish larvae. Strand-specific RT-PCR shows that (-)ss HuNoV RNA was detected intracellularly in macrophages at 2 dpi. (d) Experimental design for transcriptional profiling of FACS-sorted macrophages. (e) Violin plots show the normalized HuNoV expression per cell per macrophage subcluster. Heatmaps show the expression of genes linked to (f) phagocytosis, (g) M1 inflammatory macrophages, (h) hepatocytes, and (i) enterocytes type 2 across the macrophage subclusters. HuNoV = human norovirus, VP1 = viral protein 1, major capsid protein, dsRNA = double-stranded RNA, ss = single-stranded.
|

