Fig. 7
- ID
- ZDB-FIG-240910-43
- Publication
- García-Cuesta et al., 2024 - Allosteric modulation of the CXCR4:CXCL12 axis by targeting receptor nanoclustering via the TMV-TMVI domain
- Other Figures
- All Figure Page
- Back to All Figure Page
|
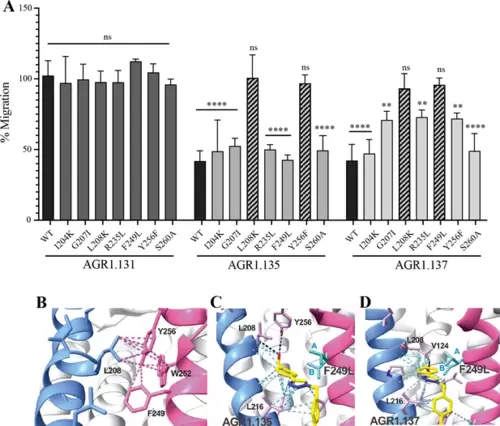
The antagonistic behavior of AGR1.135 and AGR1.137 depends on specific residues of CXCR4. (A) CXCL12-induced migration of AGR1.131- AGR1.135- or AGR1.137- pretreated JK−/− cells transiently transfected with CXCR4wt or the different mutants described. Data are shown as the mean percentage ± SD of input cells that migrate (n=4; n.s. not significant, **p≤0.01, ****p≤0.0001). (B) Cartoon representation of Y256 and its intramolecular interactions in the CXCR4 X-ray solved structure 3ODU. (C, D) Cartoon representation of the interaction of CXCR4 F249L mutant with AGR1.135 and AGR1.137, respectively. The two most probable conformations of leucine rotamers are represented in cyan. Van der Waals interactions are depicted in cyan dashed lines and hydrogen bonds in black dashed lines. |