Fig. 3
- ID
- ZDB-FIG-240910-34
- Publication
- García-Cuesta et al., 2024 - Allosteric modulation of the CXCR4:CXCL12 axis by targeting receptor nanoclustering via the TMV-TMVI domain
- Other Figures
- All Figure Page
- Back to All Figure Page
|
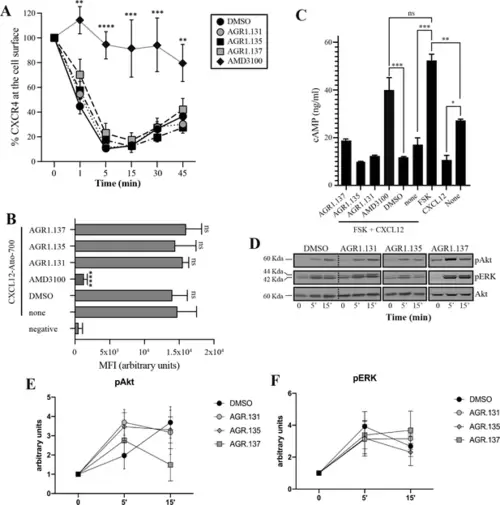
AGR1.135 and AGR1.137 treatments do not block CXCL12-mediated signaling pathways. (A) Cell surface expression of CXCR4 in Jurkat cells after stimulation with CXCL12 (12.5 nM) at different time points and analyzed by flow cytometry using an anti-CXCR4 antibody in nonpermeabilized cells. Results show mean ± SEM of the percentage of CXCR4 expression at the cell surface (n=4; n.s. not significant; **p≤0.01, ***p≤0.001, **** p≤0.0001). (B) CXCL12-Atto-700 binding on untreated or Jurkat cells pre-treated with DMSO (vehicle), AGR1.131, AGR1.135, AGR1.137 or with AMD3100 as a control, followed by flow cytometry analysis. Results are expressed as mean fluorescence intensity (MFI) values (arbitrary units). Negative corresponds to basal cell fluorescence in the absence of CXCL12-Atto-700 (mean ± SD, n=3 n.s. not significant; ****p≤0.0001). (C) Cells untreated or pretreated with the small compounds (50 μM, 30 min 37 °C) or AMD3100 (1 μM, 30 min 37 °C) were stimulated with CXCL12 (50 nM, 5 min, 37 °C) followed by forskolin (10 μM, 10 min, 37 °C). Cells were then lysed and cAMP levels were determined (mean ± SD; n=3; n.s. not significant; *p≤0.05, **p≤0.01, ***p≤0.001). (D) Western blot analysis of phospho (p) Akt and pERK in Jurkat cells pre-treated with DMSO, AGR1.131, AGR1.135 or AGR1.137, in response to CXCL12. As a loading control, membranes were re-blotted with an anti-Akt antibody. Representative experiments are shown (n=4). (E, F) Densitometry evaluation of the specific bands in D (mean ± SD; n=4). |