Fig. 5
- ID
- ZDB-FIG-240814-7
- Publication
- Emmerich et al., 2024 - Large-scale CRISPR screen reveals context-specific genetic regulation of retinal ganglion cell regeneration
- Other Figures
- All Figure Page
- Back to All Figure Page
|
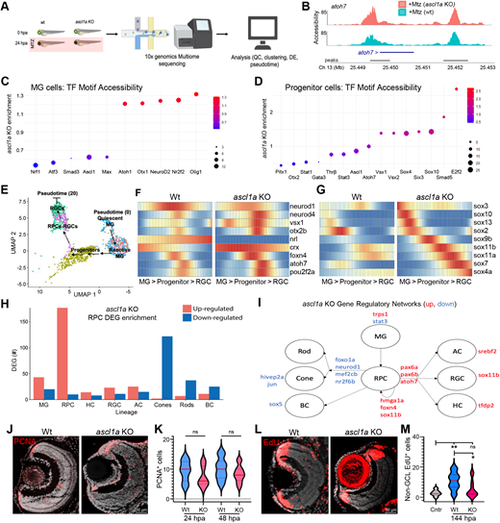
Ascl1a KO accelerates RGC regeneration kinetics by enhancing RGC fate bias. (A) Experimental setup for multiome sequencing (combined snRNA-seq and ATAC-seq) of control (wt) and ascl1a KO larvae at 0 and 24 hpa. (B) Chromatin accessibility comparison between wt and ascl1a KO larvae at the atoh7 locus after RGC ablation (+Mtz). (C) Transcription factor motif accessibility enrichment in MG cells of ascl1a KO larvae. (D) Transcription factor motif accessibility enrichment in progenitor cells of ascl1a KO larvae. (E) UMAP used to create pseudotime trajectory of MG>progenitor>RGC cells. (F,G) Heatmaps of select pro-neural (F) and Sox family (G) TF DEGs across pseudotime for wild-type and ascl1a KO larvae at 24 hpa. (H) DEGs in progenitor cells of ascl1a KO larvae associated with known retinal cell lineages. (I) Derived gene regulatory networks (GRNs) altered in ascl1a KO progenitors based on changes in expression of transcription factors known to regulate DEGs in H. (J) Representative PCNA immunostaining in RGC-ablated control (+Mtz) and RGC-ablated ascl1a crispant (+Mtz, ascl1a KO) retinas. (K) Quantification of PCNA+ cells after RGC ablation at 24 and 48 hpa in wild-type and ascl1a KO retinas. (L) Representative EdU immunostaining images of ‘secondary’ EdU lineage-traced cells (0-48 h EdU pulse, 6-day chase) in RGC-ablated wild-type and ascl1a KO retinas. (M) Quantification of the EdU+ retinal cells in the ONL and INL (excluding CMZ and GCL) in unablated controls and RGC-ablated wild-type and ascl1a KO retinas at 6 dpa. *P≤0.05, **P≤0.01. |