Fig. 4
- ID
- ZDB-FIG-240711-28
- Publication
- Choi et al., 2024 - Exposure to elevated glucocorticoid during development primes altered transcriptional responses to acute stress in adulthood
- Other Figures
- All Figure Page
- Back to All Figure Page
|
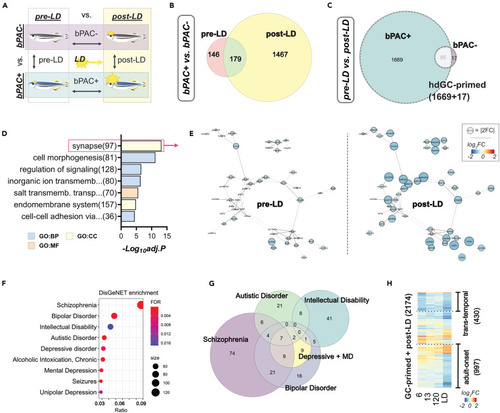
Developmental GC exposure primes differential gene expression following acute stress in bPAC+ adults (A) Schematic for three comparisons to identify transcriptional changes in response to LD exposure in adults. DEGs were determined by FC and FDR. |FC|>1.5 and FDR<0.05. (B) Venn diagram representing DEGs between bPAC+ and bPAC- in pre-LD and post-LD conditions. (C) Venn diagram showing post-LD DEGs from bPAC+ and bPAC-. The list of DEGs is described in Table S3. hdGC-primed genes are marked by a dashed line. (D) The top GO terms enriched in hdGC-primed genes. The enrichment score = -log10(adjusted p-value). (E) PPI networks of synapse-associated hdGC-primed genes. The size of the node indicates a fold-change. Networks sourced by STRING-db. (F) Result of human disorders enrichment test based on the DisGeNET database using GC-primed genes. The ratio represents overlapping/input genes. Top ranked disorders are Schizophrenia (FDR = 1.12e-04, ratio = 122/883), Bipolar Disorder (FDR = 2.47e-03, ratio = 69/477), Intellectual Disability (FDR = 1.71e-02, ratio = 61/447), Autistic Disorder (FDR = 1.87e-04, ratio = 48/261), Depressive disorder (FDR = 4.51e-03, ratio = 46/289), and Mental Depression (FDR = 4.06e-03, ratio = 42/254). The complete results are noted in Table S5. (G) Venn diagram for hdGC-primed genes that are associated with the top human psychiatric disorders in panel F. (H) Heatmap showing Log2FC of 2174 LD-DEGs across time points demonstrated 430 trans-temporal and 997 adult-onset genes were mostly downregulated. LD: looming dots, FC: fold change, FDR: False discovery rate, PPI: Protein-protein interaction, ratio: GC-primed genes/disease genes, Depressive + MD: Depressive disorder and Mental Depression. |