Fig. 2
- ID
- ZDB-FIG-240522-14
- Publication
- Madakashira et al., 2024 - DNA hypomethylation activates Cdk4/6 and Atr to induce DNA replication and cell cycle arrest to constrain liver outgrowth in zebrafish
- Other Figures
- All Figure Page
- Back to All Figure Page
|
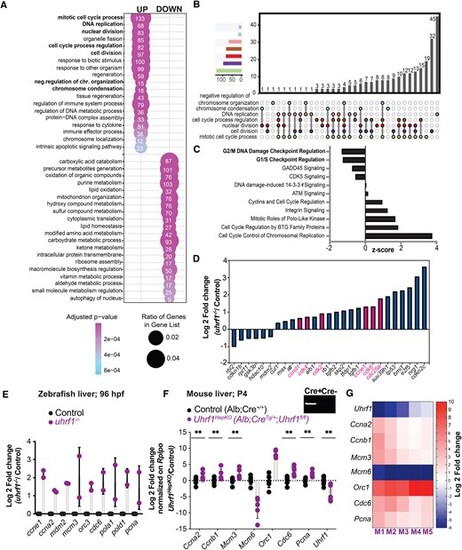
CDK4/6 is activated in uhrf1 mutant livers. (A) Gene ontology (REVIGO) of the significant upregulated and downregulated genes in 120 hpf uhrf1−/− liver RNA-Seq (P-adj < 0.05) for each category. The cell cycle GO terms are highlighted in bold. (B) UPSET plot of all the cell cycle GO category genes from A. (C) Ingenuity Pathway Analysis (IPA) of all the cell cycle genes differentially expressed in 120 hpf uhrf1−/− liver RNA-Seq, pathways involved in cell cycle checkpoint regulation in bold, z- score in IPA indicates a predicted activation or inhibition of a pathway. (D) Genes from the G1/S checkpoint pathway generated by IPA plotted with log2fold change, important cell cycle genes highlighted in red. (E) qPCR on key cell cycle genes of 96 hpf uhrf1−/− and control livers. (F) qPCR on key cell cycle genes in the livers of P4 neonatal mice deficient for Uhrf1 in hepatocytes (Uhrf1HEPKO) and controls, inset shows band for Cre transgene by RT-PCR. (G) Heatmap of Log2 fold change of key cell cycle genes and Uhrf1 in individual P4 neonatal mice livers ranked by Uhrf1 expression levels. The number of clutches (zebrafish) or individual animals (mouse) are indicated for each condition and represented as median with range. P-value *< 0.05, **< 0.005, *** < 0.0005 by unpaired Student's t-test with adjustment for multiple comparisons. |