Fig. 4
|
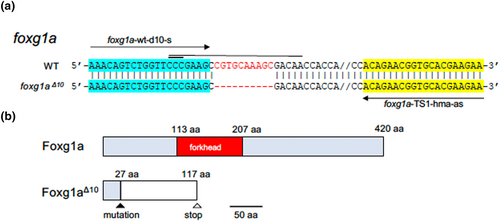
Mutations introduced into foxg1a. (a) The sequence around the site of mutation, introduced into foxg1a by the CRISPR/Cas9 method, is shown, with the wild-type sequence at the top and the mutated sequence at the bottom. Hyphens show introduced nucleotide deletions, and double dashes show the sequences omitted for convenience's sake. A horizontal thin line and thick line show the target sequence and PAM sequence, respectively. Horizontal arrows show the sequences of primers used for HMA, which are also highlighted. (b) Schematic views of the wild-type and mutant products of foxg1a. Light-blue boxes show the wild-type coding sequences, but for the functional domain that is shown in red instead, whereas the white box shows the nonsense open reading frame generated as a result of the frameshift mutation. |