Fig. 2
- ID
- ZDB-FIG-220804-13
- Publication
- Chang et al., 2022 - Zebrafish transposable elements show extensive diversification in age, genomic distribution, and developmental expression
- Other Figures
- All Figure Page
- Back to All Figure Page
|
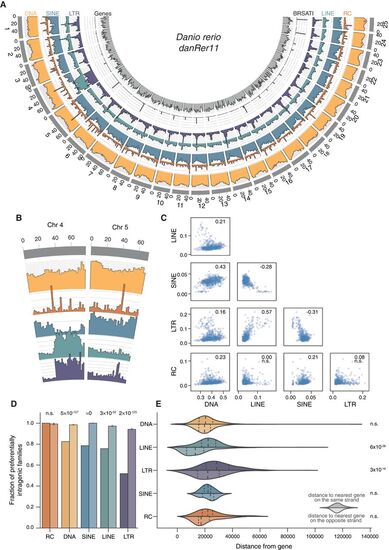
Genomic distribution of elements is nonrandom. (A) Genomic coverage of TEs in nonoverlapping 2-Mbp windows across nuclear chromosomes. Each axis line (faint gray) represents 2.5% sequence coverage. (B) Detail on Chromosomes 4 and 5. (C) Spearman's rank correlations of coverage density between major TE classes. Values for ρ given in top right corner of each plot; (n.s.) not significant. (D) TE families are defined as “preferentially intragenic” if the median distance between their insertions and the closest gene is zero; that is, most insertions in the family overlap partially or fully with gene bodies. Bars for each TE class represent observed fractions (left bars) and fractions based on random shuffling of TE insertion identities across the genome, keeping locations fixed (right bars, color desaturated). P-values calculated using binomial tests. (E) Median, per family, distance of insertions from nearest genes. Top halves indicate distance from closest gene on same strand; bottom halves (desaturated), distance from closest gene on opposite strand. P-values calculated using Wilcoxon rank-sum tests. |