Fig. 2
- ID
- ZDB-FIG-220527-31
- Publication
- Li et al., 2022 - DNA methylation safeguards the generation of hematopoietic stem and progenitor cells by repression of Notch signaling
- Other Figures
- All Figure Page
- Back to All Figure Page
|
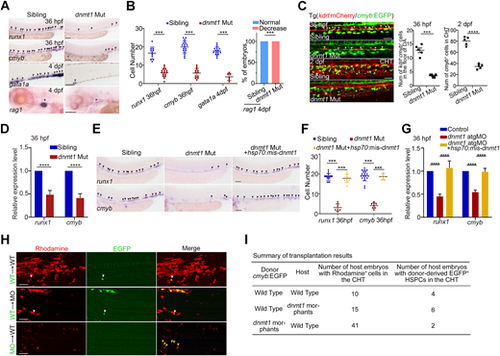
DNA methylation is required for HSPC generation. (A) Expression of HSPC markers runx1 and cmyb (arrowheads) at 36 hpf, erythroid marker gata1 (arrowheads) at 4 dpf and lymphoid marker rag1 (arrowheads) at 4 dpf in siblings and dnmt1 mutants by WISH. n≥3 replicates. (B) Quantification of WISH results in A. (C) Confocal imaging showing the number of kdrl+/cmyb+ HECs in AGM at 36 hpf (white arrowheads), and cmyb+ HSPCs in the caudal hematopoietic tissue (CHT) region at 2 dpf in siblings and dnmt1 mutants (left panels, n≥3 replicates), with quantification (right panels). n=5 embryos. (D) qPCR analysis of runx1 and cmyb expression in sibling and dnmt1 mutant embryos at 36 hpf. n=3 replicates. (E) Expression of runx1 and cmyb in sibling, dnmt1 mutant and dnmt1 mutant embryos injected with hsp70: mismatch-dnmt1-EGFP constructs. The arrowheads indicate the expression of HSPC markers runx1 and cmyb. n≥3 replicates. (F) Quantification of WISH results in E. (G) qPCR analysis of runx1 and cmyb expression in control, dnmt1 morphants and embryos co-injected with dnmt1-atgMO and hsp70: mismatch-dnmt1-EGFP constructs. n=3 replicates. (H) Transplantation results showing HSPC reconstitution in the CHT region of recipient embryos at 36 hpf. Green, cmyb+ EGFP cells; red, Rhodamine. White arrowheads show EGFP+ HSPCs contributed by donor cells; yellow arrowheads show donor-derived EGFP− hematopoietic cells in CHT region. (I) Summary of transplantation results in H. Data are mean±s.d. ***P<0.001, ****P<0.0001 (unpaired two-tailed Student's t-test). Scale bars: 100 μm (A,E); 50 μm (C,H). |