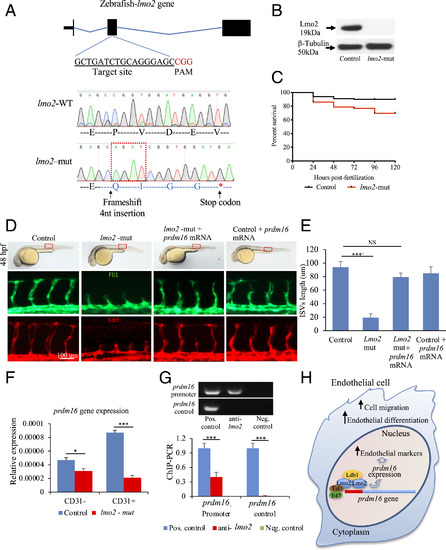
Lmo2 impacts zebrafish angiogenesis by regulating prdm16. (A) lmo2-mut in zebrafish by CRISPR/Cas9. gRNAs produced a 4-nucleotide insertion in the exon 2 and a stop codon downstream. (B) Western blot using an anti-Lmo2 antibody confirmed the absence of Lmo2. β-Tubulin was used as loading control. (C) Kaplan-Meyer survival curve of lmo2-mut zebrafish. (D) Brightfield images (Upper) showing the lmo2-mut embryo gross phenotype. Fluorescent images of the trunk region following immunostaining for Fli1 (Middle) and Kdrl (Lower), showing the reduced average length of ISVs in lmo2-mut and the rescue of vascular defects when prdm16 mRNA was injected. These data are graphically shown in E. (F) qPCR analysis showing the expression of PRDM16 in CD31+ and CD31− cell isolated from lmo2-mut embryos. (G) ChIP–PCR showing the association of Lmo2 to the prdm16 gene; gel electrophoresis of PCR products. The prdm16 promoter region, but not a control region ∼1-kb downstream, was precipitated by the anti-Lmo2 antibody. Sheared chromatin before immunoprecipitation and immunoprecipitation using spermine served as positive and negative controls, respectively. (H) Graphical abstract showing the relationship between Lmo2–prdm16 and the role of prdm16 on angiogenesis. n = 3 experiments; ANOVA test followed by Bonferroni post hoc was used to compare means. NS, not significant. ***P ≤ 0.001, *P ≤ 0.05.
|

