Fig. 8
|
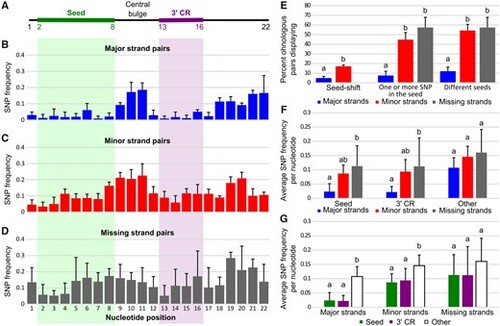
Sequence evolution of ohnologous mature miRNAs. (A) Schematic representation of a generalized 22 nucleotide long miRNA with the seed and 3′-complementary sequence regions (3′-CR) marked by green and purple bars, respectively. (B–D) Single-nucleotide polymorphism (SNP) frequencies at each nucleotide position in major, minor, and missing ohnologous strand pairs. Values represent averages across all four studied teleost species with associated SDs. (E) Frequency of major, minor, and missing nonidentical strand pairs displaying seed-shifts, SNPs in their seed, and different seeds. Values represent averages across the four studied teleost species with associated SDs. (F) SNP frequency per nucleotide in the seed, the 3′-CR, and other nucleotides (i.e., nucleotides 1, 9–12, and 17–22) of major, minor, and missing nonidentical strand pairs. Values represent averages across the four studied teleost species with associated SDs. (G) SNP frequency per nucleotide in major, minor, and missing nonidentical strand pairs for the seed, the 3′-CR, and other nucleotides. Values represent averages across the four studied teleost species with associated SDs. Different letters signify significant differences at P < 0.05. |