Figure 2
- ID
- ZDB-FIG-220310-2
- Publication
- Varga et al., 2022 - Structure-based prediction of HDAC6 substrates validated by enzymatic assay reveals determinants of promiscuity and detects new potential substrates
- Other Figures
- All Figure Page
- Back to All Figure Page
|
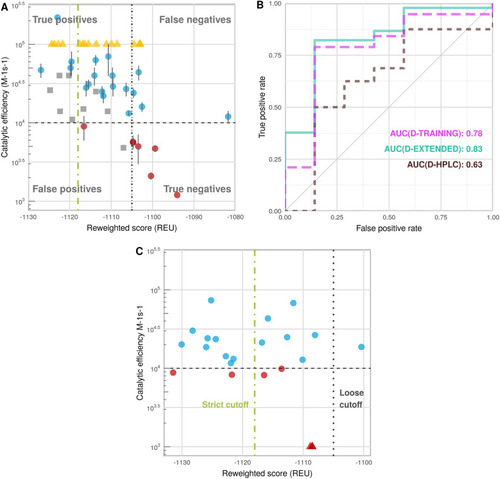
Performance of the calibrated protocol on different datasets (see Supplementary Table |