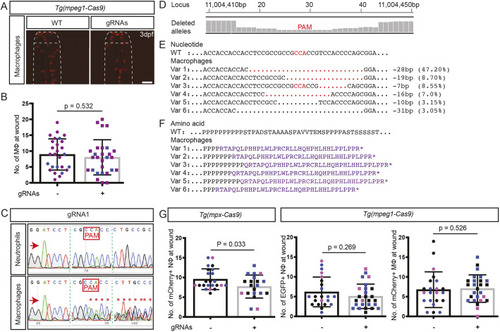
On-target trim33 gene editing in macrophages of Tg(mpeg1-Cas9) zebrafish embryos. (A) Representative fluorescent images of mCherry-labelled macrophage at caudal fin 3.5 h post-wounding in 3 dpf Tg(mpeg1-Cas9) injected with two trim33 gRNAs, compared to uninjected transgenic embryos (WT). (B) Quantification of macrophage migratory response. (C) Sanger sequencing chromatogram of an independent experiment, demonstrating on-target Cas9 activity in FACS-purified mCherry-positive macrophages. (D) Manhattan plots from NGS of the same DNA preparation as in C, displaying cumulative distribution of aligned deleted alleles at the target locus in macrophages. (E) WT reference sequence compared to six predominant variants (Var 1-6), representing 77.65% on-target gene editing in macrophages. (F) Predicted amino acid sequences of variants 1-6 with nonsense mutations. (G) Quantification of neutrophils at wound site in concurrent groups of Tg(mpx-Cas9) and Tg(mpeg1-Cas9) embryos injected with the same trim33 gRNAs, showing a neutrophil migratory defect in Tg(mpx-Cas9) neutrophils but not in Tg(mpeg1-Cas9) neutrophils. The absence of a migration defect in trim33 knockdown Tg(mpeg1-Cas9) macrophages (B) is replicated. Red arrows indicate the sequencing direction; red asterisks mark sequence heterogeneity; green vertical dashed lines indicate cropped areas of the chromatogram; PAM, protospacer adjacent motif highlighted in red boxes and font; red dots, deletion, purple font, truncated protein; black dots, sequence continues as WT. Unpaired two-tailed Student's t-test (P=0.53) of pooled data from two (B) and three (G) independent experiments indicated by different colours. Scale bar: 100 µm.
|