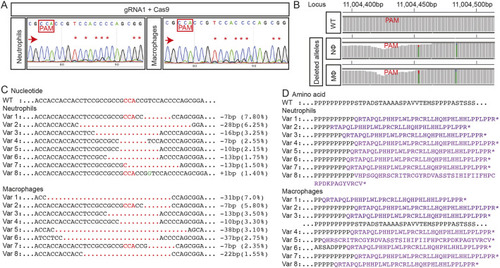
Neutrophils and macrophages from whole-body trim33 crispants have on-target gene editing. (A) Sanger chromatograms of DNA from FACS-purified neutrophils and macrophages from whole-body 3.5 dpf Tg(mpeg1:Gal4FF/UAS:NTR-mCherry)(mpx:GFP) crispant embryos injected with trim33 gRNA1 complexed with Cas9, demonstrating on-target gene editing in both leukocyte types. (B) Manhattan plots from NGS of the same DNA preparation as in A, displaying cumulative distribution of aligned deleted alleles at the target locus in neutrophils and macrophages. (C) There is a single allele in WT and eight predominant variants (Var 1-8), representing on-target gene-editing rates of 26.65% in neutrophils and 29.35% in macrophages. (D) Predicted amino acid sequences of variants 1-8; all are predicted to be nonsense mutations. Red arrows indicate the sequencing direction; red asterisks mark sequence heterogeneity; PAM, protospacer adjacent motif highlighted in red boxes and font; red dots, deletion; green font, insertion; purple font, truncated protein; black dots, sequence continues as WT.
|