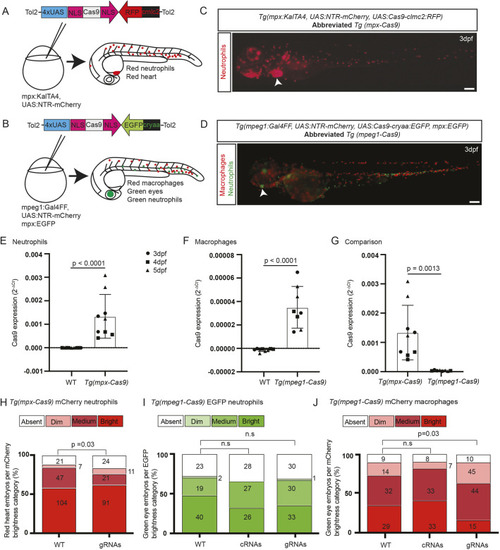
Transgenes for leukocyte lineage-specific gene editing in zebrafish. (A) For gene editing in neutrophils, a tol2-flanked UAS:Cas9 construct was microinjected into one-cell-stage Tg(mpx:KalTA4) embryos. Nuclear localization signal (NLS) motifs target Cas9 to the nucleus. This construct carried a Tg(clmc2-RFP) tracer marker. (B) For gene editing in macrophages, a different tol2-flanked UAS:Cas9 construct was microinjected into one-cell-stage Tg(mpeg1:Gal4FF) embryos that carried a Tg(cryaa-EGFP) tracer marker. Each background carried a Tg(UAS:NTR-mCherry) transgene. (C,D) Embryos with red leukocytes from the Tg(UAS:NTR-mCherry) reporter driven by either mpx:KalTA4 (neutrophil) or mpeg1:Gal4FF (macrophage) transgenes, co-segregating with the two different UAS:Cas9 transgenes indicated by their respective tracer marker [white arrowheads, red heart (C) and green eye (D)], shown for F2 Tg(mpx-Cas9) (C) and F3 Tg(mpeg1-Cas9) (D) embryos. The Tg(mpeg1-Cas9) line also carried Tg(mpx:EGFP), marking neutrophils green (D). Scale bars: 200 µm. (E-G) Quantitative PCR showing relative Cas9 mRNA expression for Tg(mpx-Cas9) (E), Tg (mpeg-Cas9) (F) and a comparison of both (G). Data normalized to housekeeping gene (ppial) and represent mean replicates of three runs at different time points differentiated by shapes: black circles, 3 dpf; black squares, 4 dpf; black triangles, 5 dpf. No differences exist between timepoints within genotypes. Unpaired two-tailed Student’s t-test. P-values are shown. (H-J) mCherry reporter gene knockdown in Tg(mpx-Cas9) and Tg(mpeg1-Cas9) embryos following delivery of a multiplexed mCherry gRNA pair targeting the transcript from the UAS:NTR-mCherry reporter (gRNA validation presented in Fig. S3). Embryos for gRNA injection were generated from Tg(mpx-Cas9) and Tg(mpeg1-Cas9) incrosses. As the incrossed fish were not confirmed as homozygous for all transgenes, there was the possibility of segregation of the Gal4 driver, UAS:Cas9 effector and UAS:NTR-mCherry reporter transgenes; hence, a categorical scoring system (absent/dim/medium/bright) was used to detect a shift towards reduced mCherry expression (further details in Fig. S3). Prior to scoring, embryos were selected for the independent backbone markers confirming the presence of the UAS:Cas9 effector [red heart for Tg(mpx-Cas9) (C, white arrowhead); green eye for Tg(mpeg1-Cas9) (D, white arrowhead)]; this line also carries the Tg(mpx:EGFP) marker resulting in green neutrophils. (H) Knockdown of mCherry expression in Tg(mpx-Cas9) neutrophils [percentage absent+dim, 15.6% (WT) versus 23.8% (gRNA injected)]. (I,J) A similar shift towards duller categories occurred in Tg(mpeg1-Cas9) mCherry macrophages (J), without any alteration in the distribution of EGFP-expressing neutrophils, serving as an internal negative control (I). Pooled data from two (H) and three (I,J) independent experiments, total pooled n values are shown within columns. P-values from chi-square test. n.s., not significant.
|