|
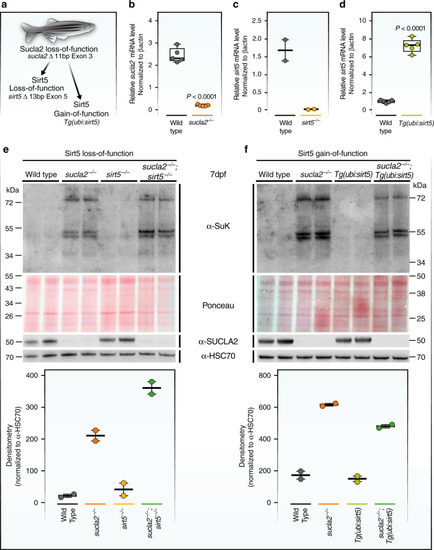
Sirt5 modulates global protein succinylation in a zebrafish model of SCL deficiency.a Schematic representation of genetic backgrounds used for zebrafish experiments. b Relative sucla2 mRNA levels in sucla2−/− zebrafish (n = 5 pools of 6 larvae, 7 dpf). c Relative sirt5 mRNA levels in sirt5−/−zebrafish (n = 2 tissue samples from adult skeletal muscle, adults of 3 months). d Relative sirt5 mRNA expression in Tg(ubi:sirt5;cryaa:zsgreen) transgenic zebrafish (n = 5 pools of 8 larvae, 8 dpf). All data in b–d, compared to wild-type siblings and normalized to βactin mRNA levels. p-values are calculated by standard unpaired t-tests for b and d. e SDS-page and western blot analysis of global protein succinylation in zebrafish larvae with homozygous mutations in sucla2, sirt5, or combined gene deficiencies and controls (7 dpf, n = 2 pools of 10–15 larvae) and f Pan-succinyl-lysine western blots of samples from larvae overexpressing sirt5 in wild-type or sucla2−/− animals (n = 2 pools of 10 to15 larvae, 7dpf). Pan-succinyl-lysine antibodies were used to quantify lysine succinylation, and Hsc70 antibodies were used as loading controls. Ponceau staining serves as an additional loading control. Boxplots in e, f show quantifications of western blots using the mean of the densitometry of the three main bands, normalized to Hsc70. The boxplots show the median, the first to third quartile, and minima and maxima. Western blot images are derived from a single experiments. Source data are provided as a Source Data file. Bp = base pairs.
|