Figure 2
- ID
- ZDB-FIG-200610-10
- Publication
- Liao et al., 2019 - RNA Granules Hitchhike on Lysosomes for Long-Distance Transport, Using Annexin A11 as a Molecular Tether
- Other Figures
- All Figure Page
- Back to All Figure Page
|
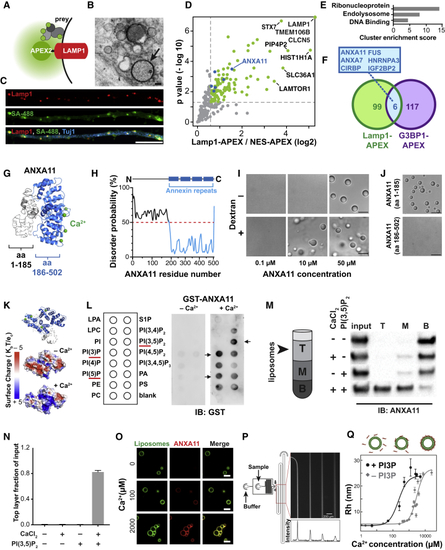
Identification of ANXA11 as a Potential Mediator of RNA Granule-Lysosome Associations (A–G) Proximity labeling proteomic screen for lysosomal interacting proteins in i3Neurons. (A) Schematic of LAMP1-APEX2 bait. (B) Electron microscopy image of DAB precipitate generated by LAMP1-APEX2 (dark contrast, arrow) surrounding lysosomes in i3Neurons. Scale bar: 100 nm. (C) Confocal immunofluorescence image of LAMP1-APEX2 biotinylated prey (streptavidin-488 staining) surrounding LAMP1-positive lysosomes in i3Neuron axons (Tuj1). Scale bar: 10 μm. (D) Plot showing statistically significant LAMP1-APEX2 enriched prey proteins from proximity-labeling proteomics in i3Neurons. n = 4, p values corrected for multiple comparisons. (E) Functional Annotation Clustering of DAVID Gene Ontology terms of Lamp1-APEX enriched prey. (F) Venn diagram of LAMP1-APEX2 hits versus G3BP1-APEX2 stress-granule hits ( (G) Predicted structural analysis of ANXA11 revealed four C-terminal calcium-binding annexin repeats (blue), and a disordered N-terminal region. (H–J) Recombinant ANXA11 undergoes liquid-liquid phase separation (H) PrDOS analysis of ANXA11 predicted a high likelihood of disorder of aa 1-185. (I) Full-length ANXA11 formed spherical, fusing liquid droplets at concentrations above 50μM (upper panel). Phase separation of ANXA11 was facilitated by 10% dextran, with phase separation occurring at lower ANXA11 concentrations (≥ 10μM). Scale bar: 5 μm. See also (J) The disordered N-terminus (aa 1-185, upper panel) of ANXA11 but not C-terminus (aa 186-502, lower panel) underwent liquid-liquid phase separation. Scale bar: 5 μm. (K–Q) Recombinant ANXA11 interacts with negatively charged lysosome-associated phospholipids. (K) Surface maps of predicted ANXA11 structure +/– Ca2+ showing increased positive surface charge (blue) in the presence of Ca2+. Top panel shows orientation of ANXA11 and location of Ca2+ ions (green). (L) Protein lipid overlay assay of recombinant ANXA11-GST protein with membrane lipids. Recombinant ANXA11-GST protein was incubated with a membrane lipid strip +/– Ca2+, followed by anti-GST immunoblotting. Arrowheads indicate enriched lipid binding. Red line highlighted the correlated phospholipid species. (M) Liposome flotation assay of recombinant ANXA11 with liposomes containing PI(3,5)P2 in the absence or presence of Ca2+. Liposomes with associated proteins floated to the top layer following ultra-centrifugation (schematic). ANXA11 in the top (T), middle (M) and bottom (B) fractions was detected via anti-ANXA11 western blot. (N) Quantification ANXA11 enrichment in the top liposome fraction in (O) Microscopy analysis of calcium-dependent recruitment of recombinant ANXA11 protein to fluorescent PI3P-containing liposomes. Representative images showed ANXA11 binding to PI3P-containing liposomes at the indicated calcium concentrations. Scale bar, 5μm. (P) Microfluidic device design for diffusional sizing assay of calcium-dependent ANXA11 binding to liposomes. Inset indicates detection area. The fluorescence intensity along the channel indicates different diffusion times of ANXA11. Scale bar, 200 μm. (Q) Microfluidic diffusional sizing assay to assess changes in molecular radius of ANXA11 upon Ca2+-dependent binding to liposomes (top panel). Bottom panel: Quantification of hydrodynamic radius of ANXA11 when binding to liposomes with (dots) and without (diamonds) PI3P versus Ca2+ concentration. Data were fitted with a Hill binding model. |
Reprinted from Cell, 179, Liao, Y.C., Fernandopulle, M.S., Wang, G., Choi, H., Hao, L., Drerup, C.M., Patel, R., Qamar, S., Nixon-Abell, J., Shen, Y., Meadows, W., Vendruscolo, M., Knowles, T.P.J., Nelson, M., Czekalska, M.A., Musteikyte, G., Gachechiladze, M.A., Stephens, C.A., Pasolli, H.A., Forrest, L.R., St George-Hyslop, P., Lippincott-Schwartz, J., Ward, M.E., RNA Granules Hitchhike on Lysosomes for Long-Distance Transport, Using Annexin A11 as a Molecular Tether, 147-164.e20, Copyright (2019) with permission from Elsevier. Full text @ Cell