Figure 3 - figure supplement 1
- ID
- ZDB-FIG-190723-951
- Publication
- Hardy et al., 2019 - Detailed analysis of chick optic fissure closure reveals Netrin-1 as an essential mediator of epithelial fusion
- Other Figures
- All Figure Page
- Back to All Figure Page
|
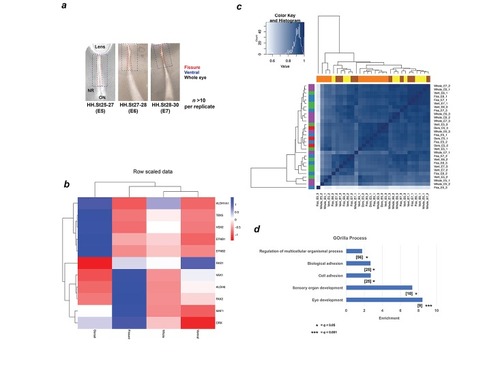
Schema and validation data for transcriptional profiling during OFC. ( |