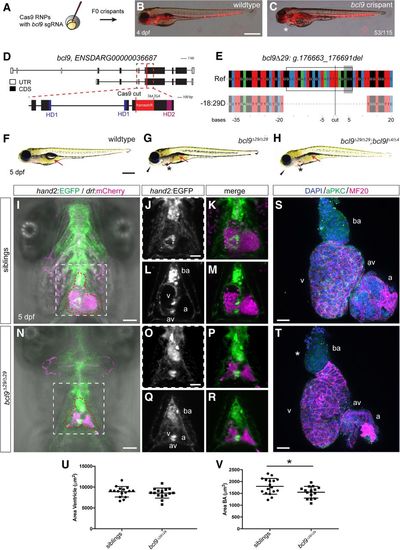
Mutations in bcl9 lead to cardiac defects in zebrafish. (A) A CRISPR–Cas9-mediated mutagenesis identified bcl9 as a potential regulator of heart morphogenesis. (B,C) bcl9 crispants have heart-looping defects, as visible in drl:mCherry transgenics, leading to cardiac edema (asterisk) (lateral view; anterior is to the left). (D) Schematic representation of the bcl9 gene locus and generation of the bcl9Δ29 germline allele. A sgRNA was designed to target the coding exon 6 between HD1 and HD2 of the zebrafish bcl9 gene. The locus is represented as per annotation Zv10, with two isoforms that differ in the first coding exon and the untranslated regions (UTRs). The dotted box represents a zoomed region of the locus, and the red line marks the location of the sgRNA to introduce the Cas9-mediated cut, which resulted in the indicated frameshift (red box) followed by two stop codons in the isolated bcl9Δ29 allele. In the isolated allele, black boxes mark coding exons (CDS), white boxes mark UTRs, blue boxes represent the CDSs that contribute to HD1, and purple boxes represent the CDSs that contribute to HD2. (E) CrispRVariants panel plot depiction of the bcl9Δ29 germline allele with a 29-base-pair (bp) deletion. The top shows genomic reference bcl9Δ29 (bcl9:g.176663_176691), shown below. bcl9Δ29 features an out-of-frame deletion introducing a frameshift followed by 157 novel amino acids terminated by two consecutive stop codons, thus disconnecting HD1 from HD2. The black box indicates the exact position of the sgRNA sequence, the gray-shaded box indicates the 5′-NGG-3′ PAM sequence, and the black line indicates the predicted Cas9-induced double-strand break position. (F–H) Bright-field images of 5-d post-fertilization (dpf) homozygous bcl9Δ29 and bcl9Δ29;bcl9lΔ4 embryos and their wild-type-looking siblings (lateral views; anterior is to the left). Mutant embryos showed heart-looping defects and cardiac edema (asterisks). Moreover, mutant embryos did not inflate their swim bladders (arrows), presumably due to a failure in gasping air because of craniofacial malformations (black arrowheads). (I–R) Single-plane illumination microscopy (SPIM) images of hand2:EGFP;drl:mCherry-expressing wild-type siblings and homozygous bcl9Δ29 embryos (ventral views; anterior is to the top; imaged after viable heart-stopping BDM treatment). I and N depict maximum-intensity projections, and J, K, O, and P show close-ups of the dotted square in I and N, while L, M, Q, and R depict optical sections at the atrio–ventricular canal level. Compared with siblings that form correctly looped hearts with atrio–ventricular canal valves and a bulbus arteriosus (BA; heart outlined with red dotted line; n = 4; J–M), bcl9Δ29 embryos show heart-looping defects (N–R) and craniofacial malformations in both cartilage and head vascularization (n = 8; N). (S–V) Six-micrometer Z-confocal projections of sibling control (S) and homozygous bcl9Δ29 (T) hearts at 5 dpf. Quantification of the ventricular (U) and BA area (V) in sibling hearts (n = 16) compared with homozygous bcl9Δ29 hearts (n = 15) shows that the BA area is significantly smaller in the bcl9Δ29 hearts. (*) P < 0.0221, unpaired t-test with Welch correction. Each data point represents the averaged ventricular or BA area from one heart; quantification was derived from three independent experiments. N = 3. (ba) BA; (a) atrium; (v) ventricle; (av) atrio–ventricular canal. Bars: B,C,F–H, 500 µm; I–R, 100 µm; S,T, 20 µm.
|

