|
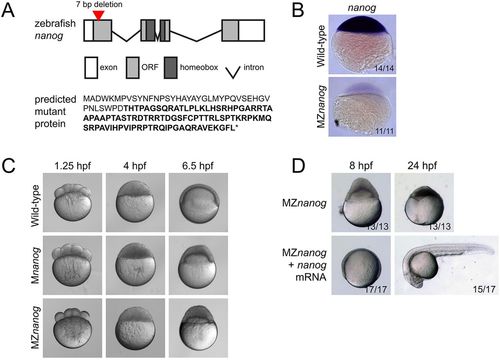
Generation and phenotype of MZnanog mutants. (A) Top: TALENs were used to generate a 7 bp deletion within the first exon of nanog. Exons, introns, the open reading frame (ORF) and the homeobox domain are indicated. Bottom: The predicted mutant protein sequence, with intact amino acids (40 of 384 total) in normal font and frameshifted amino acids in bold font. Asterisk indicates a premature stop codon. (B) In situ hybridization for nanog expression in wild-type and MZnanog embryos at sphere stage. (C) Wild-type, Mnanog and MZnanog embryos imaged at 1.25 hpf (8-cell stage), 4 hpf (sphere stage) and 6.5 hpf (shield stage in wild type). Epiboly defects in Mnanog and MZnanog are apparent at 6.5 hpf. (D) MZnanog embryos are shown at 8 hpf and at 24 hpf, either uninjected or injected with 5 pg nanog mRNA at the 1-cell stage. In B and D, the number of embryos exhibiting the illustrated phenotype is shown in the bottom-right corner of each image as a proportion of total embryos examined.
|