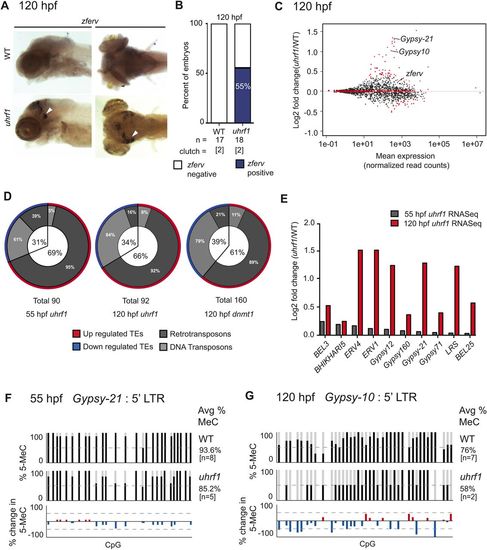
Endogenous retrotransposon expression is upregulated in uhrf1 mutants. (A) ISH of zferv expression in representative 120 hpf larvae. Staining is apparent in the eye and optic tectum (arrowheads). (B) ISH quantification of total number of embryos from two clutches scored for zferv expression in the head. (C) Expression of individual TEs, as normalized read counts, is plotted against log2 fold change of analogous TEs in uhrf1 relative to WT siblings as identified through RNA-seq. Red dots indicate TEs with significantly altered expression (P<0.05) and include the highly expressed Gypsy-10 and Gypsy-21. (D) TEs showing significant changes in C and two additional RNA-seq datasets from 55 hpf uhrf1 and 120 hpf dnmt1 mutants were grouped by expression and subdivided into transposon classes depending on their mechanism of mobility. (E) Expression comparison of select significantly induced TEs shared between 55 and 120 hpf uhrf1 mutant RNA-seq datasets. (F,G) Sanger sequencing of BS-PCR products from the Gypsy-21 (F) or Gypsy-10 (G) locus at 55 and 120 hpf in WT and uhrf1 mutants. Bottom panel shows differential methylation.
|

