Fig. 4
- ID
- ZDB-FIG-160818-24
- Publication
- Rochon et al., 2016 - Alk1 controls arterial endothelial cell migration in lumenized vessels
- Other Figures
- All Figure Page
- Back to All Figure Page
|
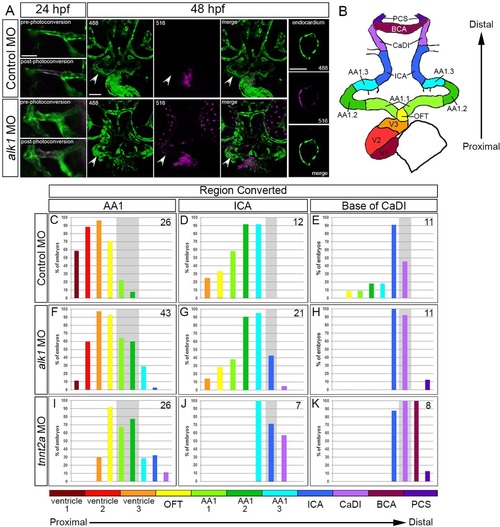
Arterial endothelial cell displacement towards the heart requires alk1 and blood flow. (A) Representative two-dimensional confocal projections of control and alk1-morphant Tg(fli1a:GAL4FF)ubs4;Tg(UAS:kaede)rk8 embryos at 24hpf (pre- and post-photoconversion of AA1 with 405nm laser) and 48hpf. Green, native Kaede; magenta, photoconverted Kaede. Arrowheads denote segment originally photoconverted. 24hpf: merge; dorsal view, anterior down. 48hpf: 488nm laser (native Kaede; green), 516nm laser (photoconverted Kaede; magenta), and merge; frontal view, dorsal up. Trio of images at right shows a single plane of a wild-type ventricle at 48hpf, demonstrating the contribution of AA1-derived cells to the endocardium. Scale bars: 50µm. (B) Schematic of 48hpf heart and cranial vasculature showing color-coded segmental boundaries used for analysis of location of photoconverted cells. (C-K) Percentage of 48hpf control (C-E), alk1-morphant (F-H) and tnnt2a-morphant (I-K) embryos exhibiting photoconverted cells in specified region of heart and cranial vessels; data are laid out proximal (left) to distal (right) with respect to the heart. Cells were photoconverted at 24hpf in proximal AA1 (C,F,I), proximal ICA (D,G,J) or at the CaDI/ICA junction (E,H,K). Gray shading denotes site of photoconversion. n (number of embryos) noted in each panel, upper right. |
| Gene: | |
|---|---|
| Fish: | |
| Knockdown Reagent: | |
| Anatomical Terms: | |
| Stage Range: | Prim-5 to Long-pec |