Fig. 5
- ID
- ZDB-FIG-140922-16
- Publication
- Sasaki et al., 2014 - Aberrant Autolysosomal Regulation Is Linked to The Induction of Embryonic Senescence: Differential Roles of Beclin 1 and p53 in Vertebrate Spns1 Deficiency
- Other Figures
- All Figure Page
- Back to All Figure Page
|
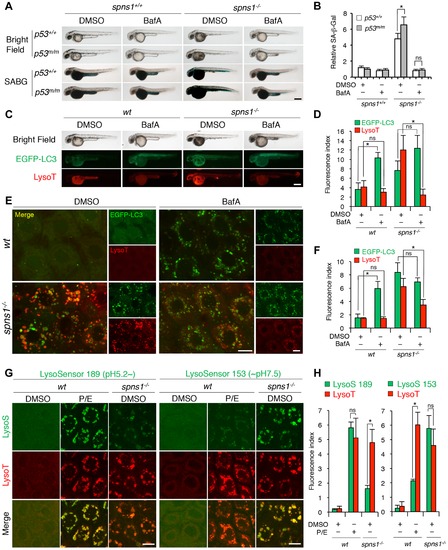
Acidity-dependent lysosomal biogenesis is rate limiting in spns1-mutant zebrafish. (A) Effect of bafilomycin A1 (BafA) on the yolk opaque phenotype (BF; bright field) and embryonic senescence (SABG; SA-β-gal) in the spns1 mutant in the presence or absence of p53 at 48 hpf. Normal wild-type (spns1+/+;p53+/+), tp53zdf1/zdf1 (p53m/m), spns1hi891/hi891 (spns1-/-) and spns1hi891/hi891;tp53zdf1/zdf1 (spns1-/-;p53m/m) embryos at 36 hpf were incubated with BafA (200 nM) for 12 h, and stained with LysoTracker at 48 hpf, followed by SA-β-gal staining at 60 hpf. Scale bar, 250 μm. (B) Quantification of the SA-β-gal intensities shown in (A). Quantification of data presented in panel A (n = 12) is shown; the number (n) of animals is for each genotype with DMSO or BafA. (C) Gross morphology, EGFP-LC3 and LysoTracker intensities in wild-type (wt) and spns1-mutant animals treated with BafA shown at 48 hpf (12 h treatment starting at 36 hpf). Scale bar, 250 μm. (D) Quantification of the EGFP-LC3 and LysoTracker fluorescence intensities shown in (C). Quantification of data presented in the middle and bottom rows (green; EGFP, red; mCherry) in panel C (n = 12) is shown; the number (n) of animals is for each genotype with DMSO or BafA. (E) Intracellular autolysosome formation and lysosomal biogenesis in the BafA-treated spns1 mutant. The samples analyzed in (C) were observed by using confocal microscopy at high magnification (×600). Scale bar, 10 μm. (F) Quantification of the EGFP-LC3 and LysoTracker fluorescence intensities shown in (E). Quantification of data presented for EGFP (green) and mCherry (red) signals in panel E (n = 6) is shown; the number (n) of animals is for each genotype with DMSO or BafA. Three independent areas (periderm or basal epidermal cells above the eye) were selected from individual animals. (G) Insufficient intracellular acidity constituent in the spns1 mutants. Using two different acidic-sensitive probes, LysoSensor 189 and neutral-sensitive LysoSensor 153 (green), in combination with LysoTracker (red), wt and spns1-mutant animals showed detectable signals when stained at 72 hpf. In spns1-mutant animals, autolysosomal and/or lysosomal compartments were more prominently detectable by LysoSensor 153 than by LysoSensor 189, at the cellular level with enhanced signal intensity of these enlarged compartments. In stark contrast, the cellular compartments in wt fish treated with pepstatin A and E-64-d (P/E) (12 h treatment from 60 hpf through 72 hpf) were more prominently detectable by LysoSensor 189 than by LysoSensor 153 under the identical LysoTracker staining conditions. Of note, these autolysosomal and lysosomal compartments in spns1 mutants, as well as in wt animals treated with pepstatin A and E-64-d, may still retain some weak (higher pH) and strong (lower pH) acidity, respectively, as short-term BafA treatment (for 1 h between 71 and 72 hpf) can abolish the acidic compartments stained by both LysoSensor and LysoTracker (Figure S17C and D). Scale bar, 10 μm. (H) Quantification of the LysoSensor (189 and 153) and LysoTracker fluorescence intensities shown in (G). Quantification of data presented for LysoSensor (green) and LysoTracker (red) signals in panel G (n = 6) is shown; the number (n) of animals is for each genotype with DMSO or pepstatin A and E-64-d (P/E). Three independent areas (periderm or basal epidermal cells above the eye) were selected from individual animals. Error bars represent the mean ± S.D., *p<0.005; ns, not significant. |
| Gene: | |
|---|---|
| Fish: | |
| Condition: | |
| Anatomical Terms: | |
| Stage: | Long-pec |
| Fish: | |
|---|---|
| Condition: | |
| Observed In: | |
| Stage: | Long-pec |