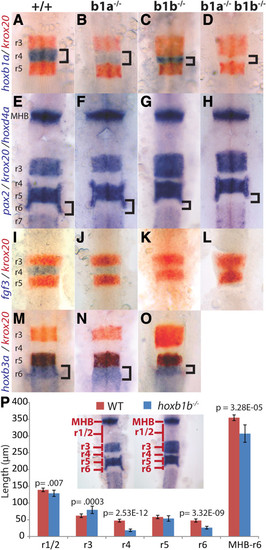
Zebrafish hoxb1b is required for hindbrain segmentation. A-O. Wild type (A, E, I, M), hoxb1a-/-(B, F, J, N), hoxb1b-/-(C, G, K, O) or doubly hoxb1a-/-;hoxb1b-/- embryos (D, H, L) were assayed by in situ hybridization for expression of hoxb1a in r4 (blue stain in A-D), krox20 in r3/r5 (red stain in A-D, I-O and blue stain in panels E-H), pax2 at the midbrain-hindbrain boundary (blue stain in E-H), fgf3 in r4 (blue stain in I-L), hoxd4a in r7 (blue stain in E-H) and hoxb3a in r5/r6 (blue stain in M-O). P. Quantification of segmentation defects in hoxb1b-/- embryos. Rhombomere lengths were measured as indicated in the inset. MHB-r6 measures the full distance from the anterior limit of the MHB to the posterior limit of r6. p-values were computed using Students’ t-test and error bars represent standard error. N = 10 embryos. All embryos are flat mounted in dorsal view with anterior to the top. A-H and M-O are at 22hpf, while I-L are at 14hpf. r = rhombomere; MHB = midbrain/hindbrain boundary.
|

