Fig. 7
- ID
- ZDB-FIG-140822-45
- Publication
- Stegmaier et al., 2014 - Fast segmentation of stained nuclei in terabyte-scale, time resolved 3D microscopy image stacks
- Other Figures
- All Figure Page
- Back to All Figure Page
|
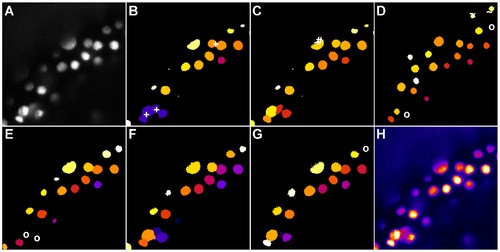
Comparison of the segmentation quality achieved by the investigated algorithms on a 3D image of labeled nuclei of a zebrafish embryo acquired using DSLM. The panels show the maximum intensity projection of 3 neighbouring z-slices. Original image (A), adaptive thresholding using Otsu’s method [23] (B), Otsu’s method combined with watershed-based blob splitting [23], [30] (C), geodesic active contours [31] (D), gradient vector flow tracking [16] (E), graph-cuts segmentation [17] (F), TWANG segmentation (G) and a false colored original image (H). The symbols indicate segmentation errors for nuclei that are either split (#), merged (+), missing (o) or spurious (~). |