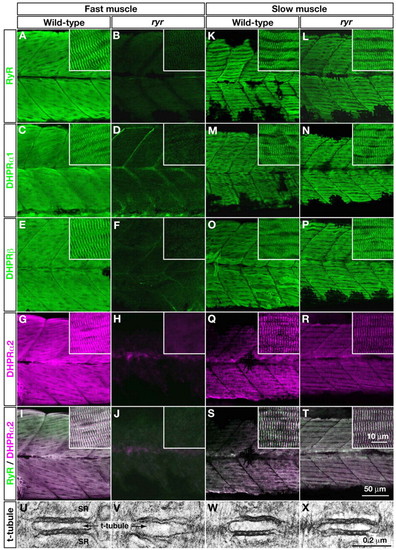
E-C-coupling components are dramatically decreased in ryr fast muscles. (A-H,K-R) The distribution of RyRs and DHPRs were assayed in fast and slow muscles at 48 hpf with anti-RyR (A,B,K,L), anti-DHPRα1 (C,D,M,N), anti-DHPRß (E,F,O,P) and anti-α2 (G,H,Q,R). Wild-type fast muscles express RyR (A), DHPRα1 (C), DHPRß (E) and DHPRα2 (G) in a striated pattern that presumably corresponds to the t-tubule-SR junctions, whereas clustering is not observed in mutant fast muscles (B,D,F,H). Wild-type slow muscles express RyR (K), DHPRα1 (M), DHPRß (O) and DHPRα2 (Q) in a striated pattern, as in wild-type fast muscles. ryr mutant slow muscles also express RyR (L), DHPRα1 (N), DHPRß (P) and DHPRα2 (R) in a striated pattern. (I,J,S,T) RyR (green) and DHPRα2 (purple) are colocalized in wild-type fast (I) and slow (S) muscle and in mutant slow muscle (T) but not in ryr mutant fast muscle (J). Insets show a higher magnification of muscle fibers. (U-X) Electron micrographs of t-tubule-SR junctions in muscles at 48 hpf. Putative RyR-DHPR aggregates are visible as dense particles between t-tubule and SR membranes in wild-type fast muscle (U) but not in ryr mutant fast muscle (V). The RyR-DHPR aggregates are present in both wild-type (W) and mutant (X) slow muscles. SR, sarcoplasmic reticulum.
|

