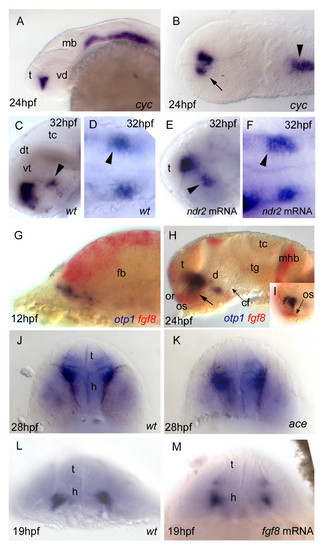
Nodal and FGF signalings are related with otp1 in the diencephalon. Anterior is left in all but J-M panels. J-M panels are frontal views, B, D and F are dorsal, and the remaining ones are lateral. A, B, Nodal signals emanating from mesendoderm and overlaying neuroectoderm represent one of the most important inducing pathways that govern regional patterning of the neural tube. Zebrafish cyclops (cyc) mutant embryos lack a functional floor plate, and indirectly disrupt HH signaling [55, 56, 57, 58]. At 24 hpf, cyc mutants display complete loss of otp1 expression in the PT, and show an asymmetric reduction of PO domains (arrow). Forebrain and hindbrain patterns are closer or fused along the midline (arrowhead). C-F, Accordingly, the essential role of Nodal signaling in otp1 transcription in the posterior hypothalamus is confirmed by microinjection of ndr2 mRNA whereas, compared with controls (C, D), otp1 domain in the PT is expanded (E, F) (arrowhead in C-F indicates the PT-specific otp1 expression) (32 hpf). G-I, Double WISH reveals that fgf8 (red) and otp1 (blue) expression patterns are complementary at the boundary between prospective telencephalon and diencephalon since early somitogenesis stages (G, 5 s).H, At 24 hpf, PO otp1 expression is adjacent to fgf8-positive optic stalks (arrow). I, A high power image of H illustrates the physical proximity of otp1 and fgf8 transcripts. J, K, fgf8 gene activity is abolished in acerebellar (ace) mutant [37]. Examination of an optic section of ace embryos at 28 hpf through the optic area shows that the PO-specific otp1 cluster is expanded radially (K) in comparison with controls (J). L, M, Overexpression of fgf8 induces a reduction of the number of otp1 expressing cells in the PO, whereas this cluster also fails to properly coalesce, likely due to developmental delay (19 hpf). On the contrary, the PT-specific otp1 domain is unaltered (data not shown). cf, cephalic flexure; d, diencephalon; dt, dorsal thalamus; fb, forebrain; h, hypothalamus; mb, midbrain; mhb, midbrain-hindbrain boundary; or, optic recess; os, optic stalks; t, telencephalon; tc, tectum; tg, tegmentum; vd, ventral diencephalon; vt, ventral thalamus.
|

