- Title
-
In vivo screening for toxicity-modulating drug interactions identifies antagonism that protects against ototoxicity in zebrafish
- Authors
- Bustad, E., Mudrock, E., Nilles, E.M., Mcquate, A., Bergado, M., Gu, A., Galitan, L., Gleason, N., Ou, H.C., Raible, D.W., Hernandez, R.E., Ma, S.
- Source
- Full text @ Front Pharmacol
|
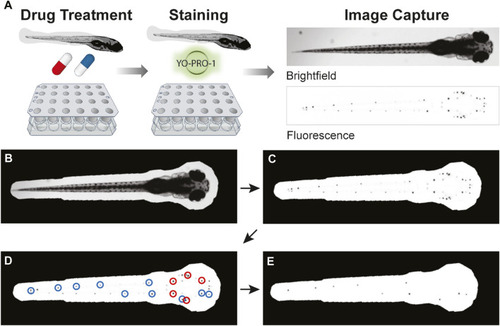
Overview of PEPITA workflow. |
|
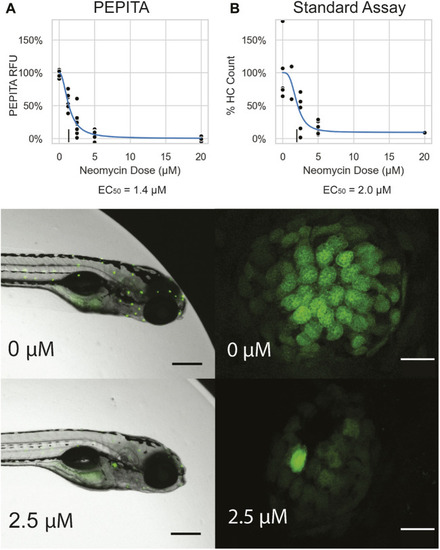
Characterization of single drug dose response with PEPITA. |
|
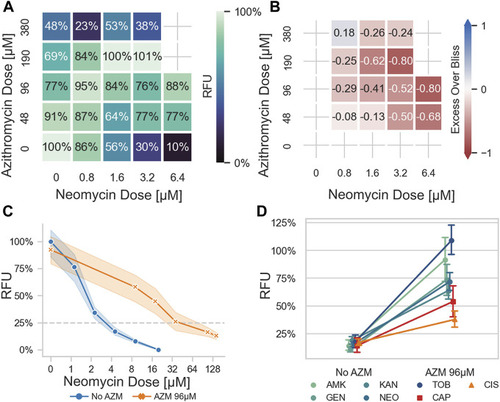
Azithromycin antagonizes aminoglycoside-induced ototoxicity in a zebrafish model. |
|
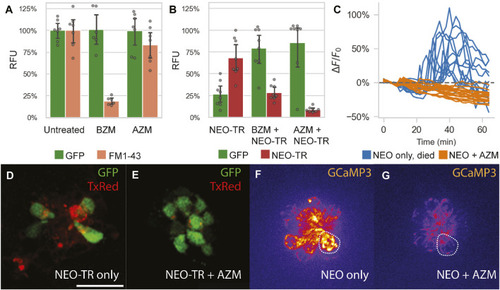
Impact of azithromycin and neomycin treatment on hair cell function. |
|
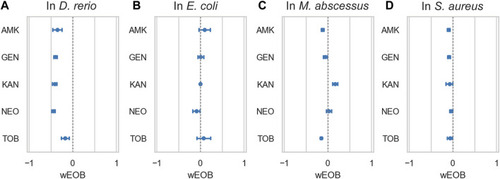
Azithromycin and aminoglycosides do not interact antagonistically in bacteria. |