- Title
-
Variants in the WDR44 WD40-repeat domain cause a spectrum of ciliopathy by impairing ciliogenesis initiation
- Authors
- Accogli, A., Shakya, S., Yang, T., Insinna, C., Kim, S.Y., Bell, D., Butov, K.R., Severino, M., Niceta, M., Scala, M., Lee, H.S., Yoo, T., Stauffer, J., Zhao, H., Fiorillo, C., Pedemonte, M., Diana, M.C., Baldassari, S., Zakharova, V., Shcherbina, A., Rodina, Y., Fagerberg, C., Roos, L.S., Wierzba, J., Dobosz, A., Gerard, A., Potocki, L., Rosenfeld, J.A., Lalani, S.R., Scott, T.M., Scott, D., Azamian, M.S., Louie, R., Moore, H.W., Champaigne, N.L., Hollingsworth, G., Torella, A., Nigro, V., Ploski, R., Salpietro, V., Zara, F., Pizzi, S., Chillemi, G., Ognibene, M., Cooney, E., Do, J., Linnemann, A., Larsen, M.J., Specht, S., Walters, K.J., Choi, H.J., Choi, M., Tartaglia, M., Youkharibache, P., Chae, J.H., Capra, V., Park, S.G., Westlake, C.J.
- Source
- Full text @ Nat. Commun.
|
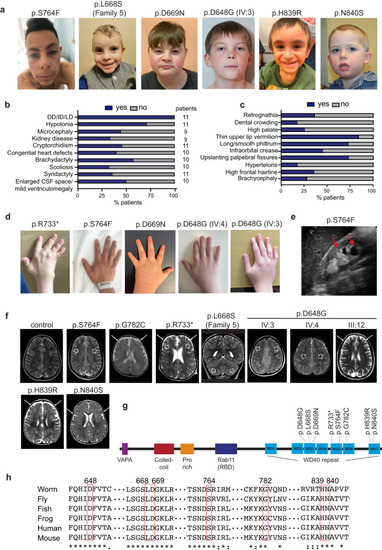
Clinical assessment of WDR44 variants. |
|
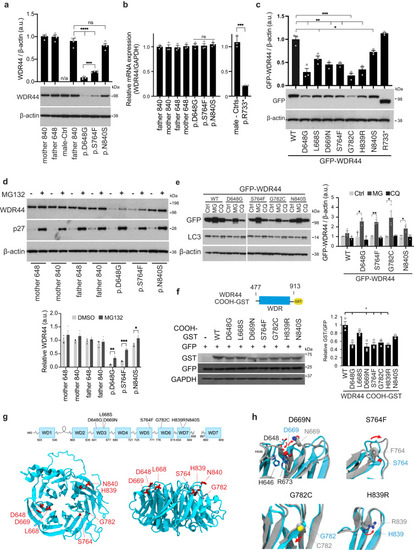
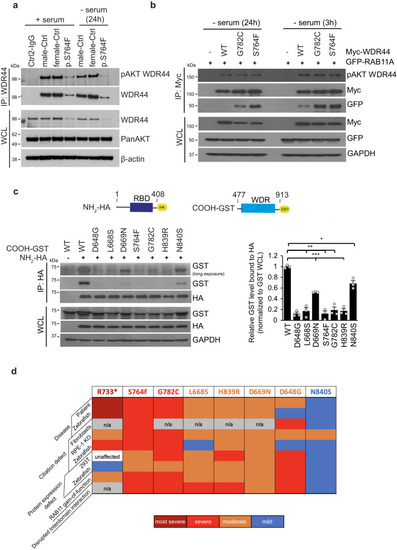
WDR44 variants affect protein stability and reduce expression. |
|
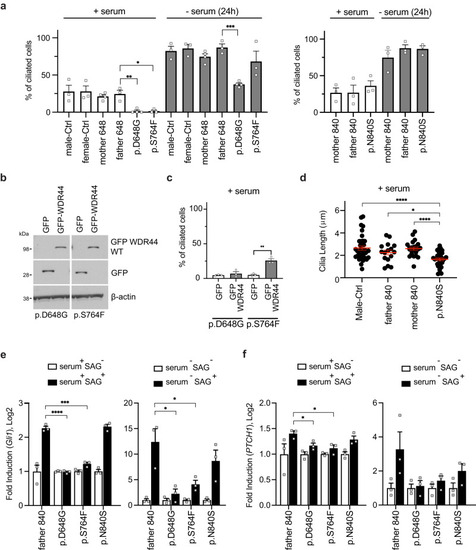
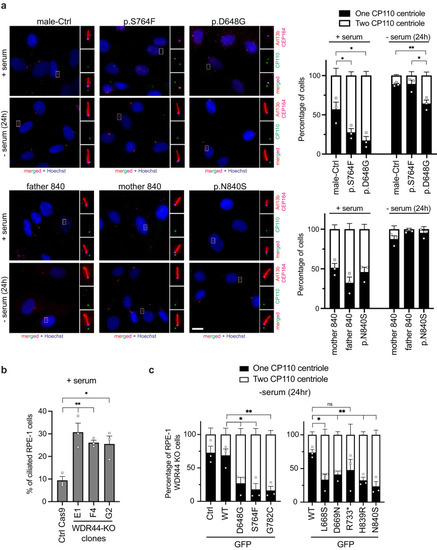
Patient fibroblasts have affected ciliation and Hedgehog signaling. |
|
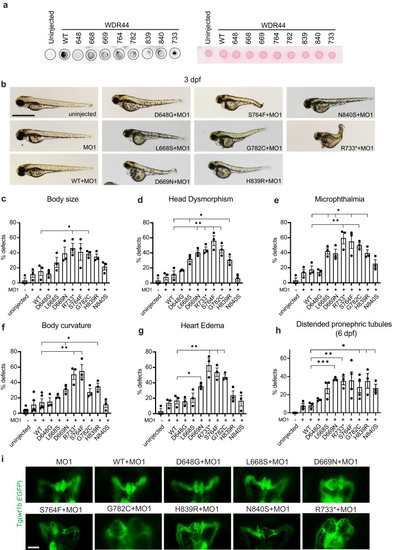
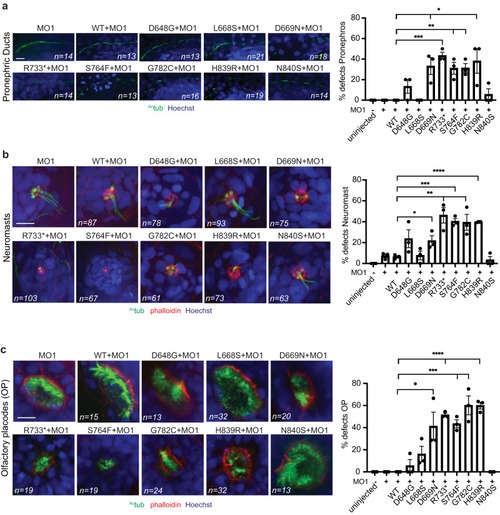
WDR44 variants affect zebrafish embryonic development. |
|
WDR44 variants reduce ciliogenesis in zebrafish embryos. Quantification ( |
|
WDR44 variants reduce ciliogenesis initiation. |
|
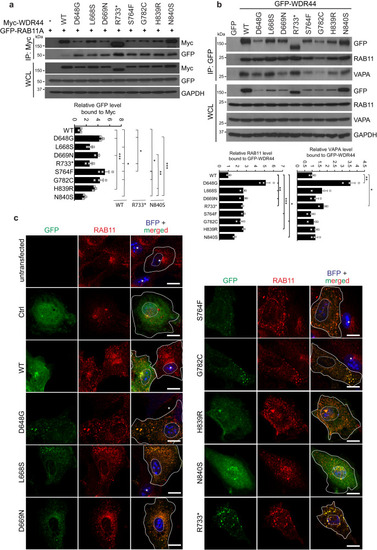
WDR44 patient variants enhance binding to RAB11 and cause RAB11 redistribution in cells. |
|
Interactions between NH2-terminus containing the RBD and the WDR are affected by patient variants. |

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. PHENOTYPE:
|