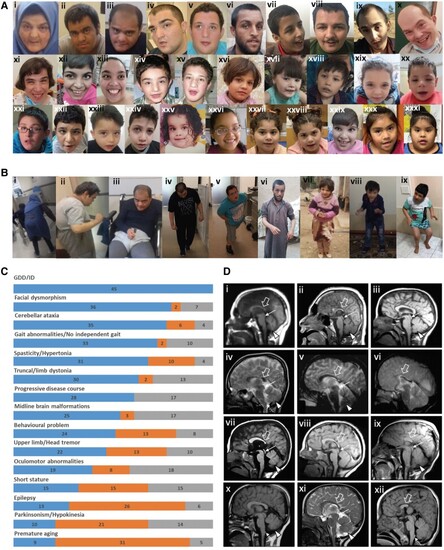
Clinical features of the affected individuals with bi-allelic ACBD6 variants. [A(i–xiii)] Representative photographs demonstrating facial features of the affected individuals in adulthood: F1:S1 (i); F1:S2 (ii); F1:S3 (iii); F3:S2 (iv); F3:S1 (v); F6:S1 (vi); F7:S2 (vii); F7:S1 (viii); F11:S1 (ix); F17:S1 (x); F17:S2 (xi); F20:S1 (xii); and F16:S1 (xiii). [A(xiv–xxxi)] Representative photographs demonstrating facial features of the affected individuals in childhood: F3:S2 (xiv) at 5 years old; F3:S1 (xv) at 12 years old; F6:S2 (xvi); F8:S1 (xvii); F8:S2 (xviii); F10:S1 (xix); F10:S2 (xx); and F11:S1 (xxi) at younger age; F13:S1 (xxii); F14:S1 (xxiii); F15:S1 (xxiv); F16:S1 (xxv) at 2 years old; F16:S1(xxvi) at 4 years old; F19:S1 (xxvii); F19:S2 (xxviii); F20:S2 (xxix); F12:S1 (xxx); and F12:S2 (xxxi). The most frequently seen facial dysmorphologies in adults are high nasal ridge, full nasal tip, small mouth, thin upper lip and broad chin. The most frequently seen facial dysmorphologies in children are bifrontal/bitemporal narrowing, arched eyebrows, hypertelorism, up-slanting palpebral fissures, depressed nasal bridge, full nasal tip, thin upper lip, full lower lip and broad chin. [B(i–ix)] Representative photographs demonstrating postural abnormalities seen in the affected individuals. A stooped body posture and lateral flexion of the trunk can be seen in the individuals F1:S1 (i), F1:S2 (ii), F1:S3 (iii), F3:S1 (iv), F3:S2 (v), F6:S1 (vi), F6:S2 (vii), F8:S2 (viii) and F13:S1 (ix). (C) Bar graph summarizing proportions of various clinical findings in the ACBD6 cohort. Blue = affected; orange = unaffected; grey = not ascertained/not applicable. GDD = global developmental delay; ID = intellectual disability. [D(i–xii)] Representative neuroimaging features of the affected individuals. Brain MRI, midline sagittal images of the affected individuals F1:S1 (i), F1:S2 (ii), F5:S1(iii), F56:S2 (iv), F7:S1 (v), F7:S2 (vi), F10:S1 (vii), F10:S2 (viii), F11:S1 (ix), F13:S1 (x), F14:S1 (xi) and F19:S1 (xii). Most of the affected individuals have corpus callosum agenesis or hypoplasia with prevalent involvement of the posterior sections (empty arrows), variably associated with short midbrain (thin arrows) and small inferior cerebellar vermis (arrowheads). In addition, mild hypertrophy of the clava was noted in some subjects (dotted arrows). Note that the anterior commissure was markedly hypoplastic or absent in all affected individuals.
|