- Title
-
Spinal cord repair is modulated by the neurogenic factor Hb-egf under direction of a regeneration-associated enhancer
- Authors
- Cigliola, V., Shoffner, A., Lee, N., Ou, J., Gonzalez, T.J., Hoque, J., Becker, C.J., Han, Y., Shen, G., Faw, T.D., Abd-El-Barr, M.M., Varghese, S., Asokan, A., Poss, K.D.
- Source
- Full text @ Nat. Commun.
|
|
|
|
|
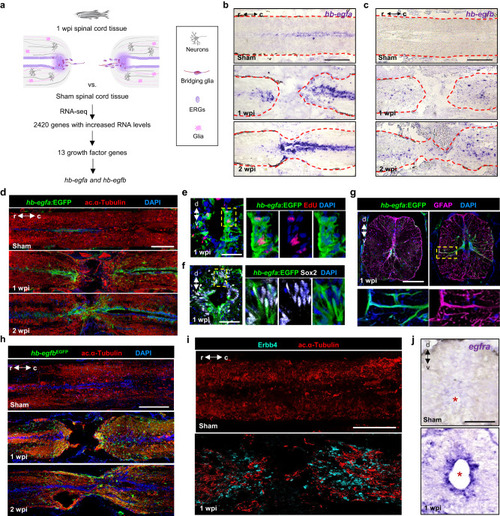
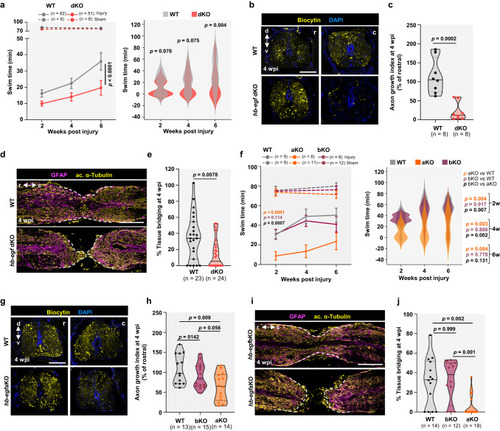
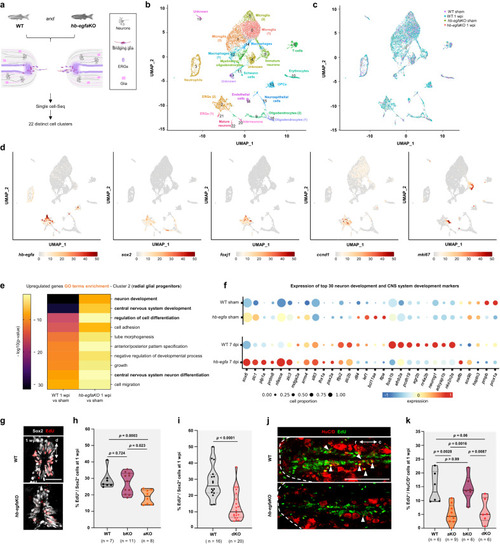
scRNA-seq identifies a role for Hb-egfa in stump neurogenesis during spinal cord repair. |
|
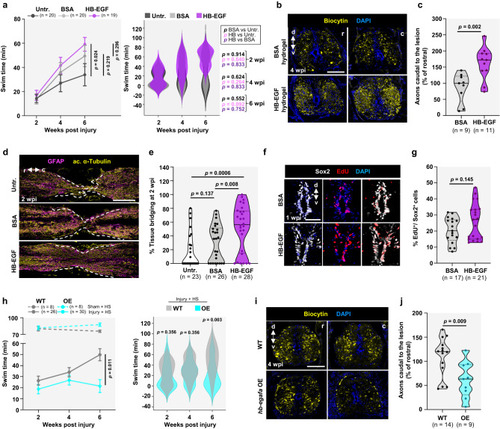
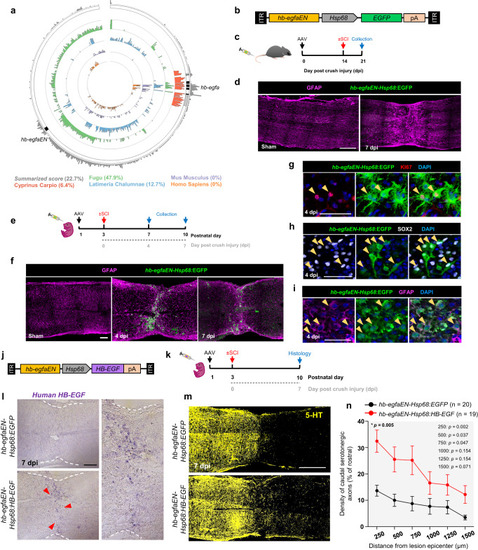
Effects of Hb-egf supplementation on spinal cord regeneration. |
|
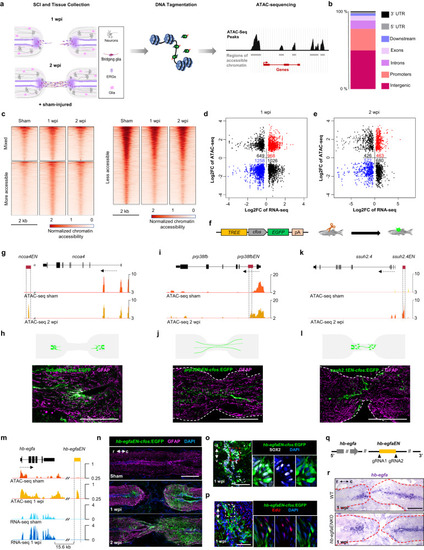
Chromatin profiling reveals an enhancer linked to |
|
|