- Title
-
Deficiency in hereditary hemorrhagic telangiectasia-associated Endoglin elicits hypoxia-driven heart failure in zebrafish
- Authors
- Lelièvre, E., Bureau, C., Bordat, Y., Frétaud, M., Langevin, C., Jopling, C., Kissa, K.
- Source
- Full text @ Dis. Model. Mech.
|
|
|
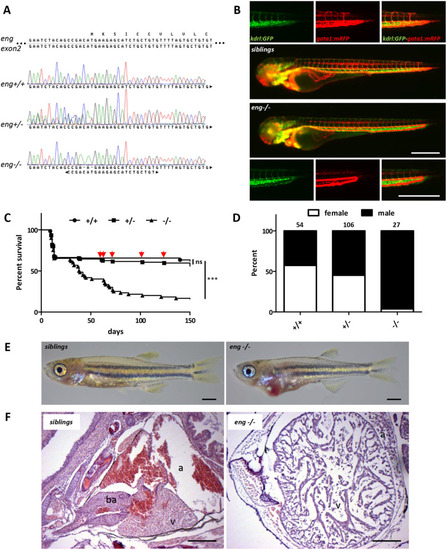
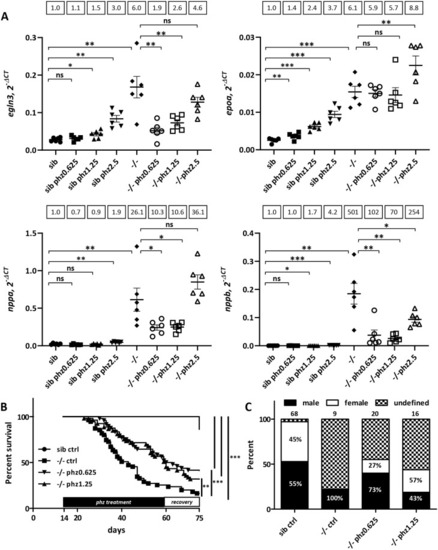
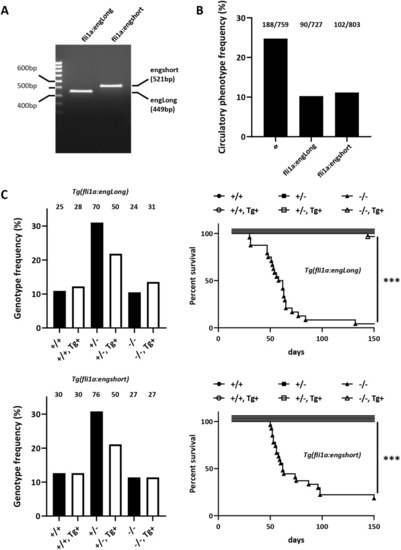
PHENOTYPE:
|
|
EXPRESSION / LABELING:
PHENOTYPE:
|
|
PHENOTYPE:
|
|
EXPRESSION / LABELING:
PHENOTYPE:
|
|
PHENOTYPE:
|